Role of the herpes simplex virus helicase-primase complex during adeno-associated virus DNA replication
- PMID: 16699004
- PMCID: PMC1472166
- DOI: 10.1128/JVI.02718-05
Role of the herpes simplex virus helicase-primase complex during adeno-associated virus DNA replication
Abstract
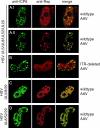
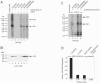
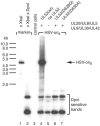
A subset of DNA replication proteins of herpes simplex virus (HSV) comprising the single-strand DNA-binding protein, ICP8 (UL29), and the helicase-primase complex (UL5, UL8, and UL52 proteins) has previously been shown to be sufficient for the replication of adeno-associated virus (AAV). We recently demonstrated complex formation between ICP8, AAV Rep78, and the single-stranded DNA AAV genome, both in vitro and in the nuclear HSV replication domains of coinfected cells. In this study the functional role(s) of HSV helicase and primase during AAV DNA replication were analyzed. To differentiate between their necessity as structural components of the HSV replication complex or as active enzymes, point mutations within the helicase and primase catalytic domains were analyzed. In two complementary approaches the remaining HSV helper functions were either provided by infection with HSV mutants or by plasmid transfection. We show here that upon cotransfection of the minimal four HSV proteins (i.e., the four proteins constituting the minimal requirements for basal AAV replication), UL52 primase catalytic activity was not required for AAV DNA replication. In contrast, UL5 helicase activity was necessary for fully efficient replication. Confocal microscopy confirmed that all mutants retained the ability to support formation of ICP8-positive nuclear replication foci, to which AAV Rep78 colocalized in a manner strictly dependent on the presence of AAV single-stranded DNA (ssDNA). The data indicate that recruitment of AAV Rep78 and ssDNA to nuclear replication sites by the four HSV helper proteins is maintained in the absence of catalytic primase or helicase activities and suggest an involvement of the HSV UL5 helicase activity during AAV DNA replication.
Figures



References
-
- Biswas, N., and S. K. Weller. 1999. A mutation in the C-terminal putative Zn2+ finger motif of UL52 severely affects the biochemical activities of the HSV-1 helicase-primase subcomplex. J. Biol. Chem. 274:8068-8076. - PubMed
-
- Biswas, N., and S. K. Weller. 2001. The UL5 and UL52 subunits of the herpes simplex virus type 1 helicase-primase subcomplex exhibit a complex interdependence for DNA binding. J. Biol. Chem. 276:17610-17619. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

