Prediction and identification of herpes simplex virus 1-encoded microRNAs
- PMID: 16699030
- PMCID: PMC1472173
- DOI: 10.1128/JVI.00200-06
Prediction and identification of herpes simplex virus 1-encoded microRNAs
Abstract
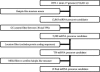
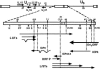
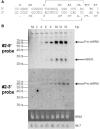
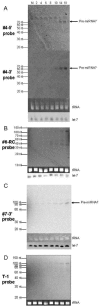
MicroRNAs (miRNAs) are key regulators of gene expression in higher eukaryotes. Recently, miRNAs have been identified from viruses with double-stranded DNA genomes. To attempt to identify miRNAs encoded by herpes simplex virus 1 (HSV-1), we applied a computational method to screen the complete genome of HSV-1 for sequences that adopt an extended stem-loop structure and display a pattern of nucleotide divergence characteristic of known miRNAs. Using this method, we identified 11 HSV-1 genomic loci predicted to encode 13 miRNA precursors and 24 miRNA candidates. Eight of the HSV-1 miRNA candidates were predicted to be conserved in HSV-2. The precursor and the mature form of one HSV-1 miRNA candidate, which is encoded approximately 450 bp upstream of the transcription start site of the latency-associated transcript (LAT), were detected during infection of Vero cells by Northern blot hybridization. These RNAs, which behave as late gene products, are not predicted to be conserved in HSV-2. Additionally, small RNAs, including some that are roughly the expected size of precursor miRNAs, were detected using probes for miRNA candidates derived from sequences encoding the 8.3-kilobase LAT, from sequences complementary to U(L)15 mRNA, and from the region between ICP4 and U(S)1. However, no species the size of typical mature miRNAs were detected using these probes. Three of these latter miRNA candidates were predicted to be conserved in HSV-2. Thus, HSV-1 encodes at least one miRNA. We hypothesize that HSV-1 miRNAs regulate viral and host gene expression.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

