MOZ is essential for maintenance of hematopoietic stem cells
- PMID: 16702405
- PMCID: PMC1472906
- DOI: 10.1101/gad.1393106
MOZ is essential for maintenance of hematopoietic stem cells
Abstract
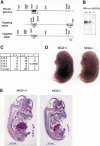
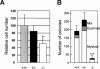
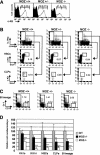
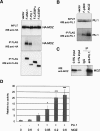
Monocytic leukemia zinc-finger protein (MOZ), a MYST family histone acetyltransferase, is involved in the chromosome translocations associated with acute myeloid leukemia. MOZ acts as a transcriptional coactivator for AML1, which is essential for establishment of definitive hematopoiesis. To investigate the roles of MOZ in normal hematopoiesis, we generated MOZ-null mice. MOZ-/- mice died around embryonic day 15 (E15). In MOZ-/- E14.5 embryos, hematopoietic stem cells, lineage-committed progenitors, and B lineage cells were severely reduced. On the other hand, arrest of erythroid maturation and elevated myeloid lineage populations were observed. MOZ-deficient fetal liver cells could not reconstitute hematopoiesis of recipients after transplantation. Analysis using microarray and flow cytometry revealed that expression of thrombopoietin receptor (c-Mpl), HoxA9, and c-Kit was down-regulated. These results show that MOZ is required for maintenance of hematopoietic stem cells, and that it plays a role in differentiation of erythroid and myeloid cells. Some aspects of the MOZ-/- phenotype are similar to that observed in PU.1-deficient mice. MOZ was able to interact with PU.1 and activate PU.1-dependent transcription, thus suggesting a physical and functional link between PU.1 and MOZ.
Figures



References
-
- Antonchuk J., Hyland C.D., Hilton D.J., Alexander W.S. Synergistic effects on erythropoiesis, thrombopoiesis, and stem cell competitiveness in mice deficient in thrombopoietin and steel factor receptors. Blood. 2004;104:1306–1313. - PubMed
-
- Borrow J., Stanton V.P., Jr., Andresen J.M., Becher R., Behm F.G., Chaganti R.S., Civin C.I., Disteche C., Dube I., Frischauf A.M., et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat. Genet. 1996;14:33–41. - PubMed
-
- Calvo K.R., Sykes D.B., Pasillas M., Kamps M.P. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression. Mol. Cell. Biol. 2000;20:3274–3285. - PMC - PubMed
-
- Carapeti M., Aguiar R.C., Goldman J.M., Cross N.C. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–3133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
