Toward a molecular understanding of pleiotropy
- PMID: 16702416
- PMCID: PMC1569710
- DOI: 10.1534/genetics.106.060269
Toward a molecular understanding of pleiotropy
Abstract
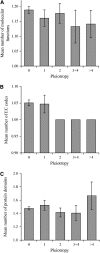
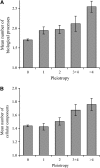
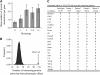
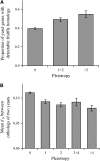
Pleiotropy refers to the observation of a single gene influencing multiple phenotypic traits. Although pleiotropy is a common phenomenon with broad implications, its molecular basis is unclear. Using functional genomic data of the yeast Saccharomyces cerevisiae, here we show that, compared with genes of low pleiotropy, highly pleiotropic genes participate in more biological processes through distribution of the protein products in more cellular components and involvement in more protein-protein interactions. However, the two groups of genes do not differ in the number of molecular functions or the number of protein domains per gene. Thus, pleiotropy is generally caused by a single molecular function involved in multiple biological processes. We also provide genomewide evidence that the evolutionary conservation of genes and gene sequences positively correlates with the level of gene pleiotropy.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

