Canine coronavirus highly pathogenic for dogs
- PMID: 16704791
- PMCID: PMC3291441
- DOI: 10.3201/eid1203.050839
Canine coronavirus highly pathogenic for dogs
Abstract
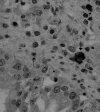
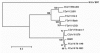
Canine coronavirus (CCoV) is usually responsible for mild, self-limiting infections restricted to the enteric tract. We report an outbreak of fatal disease in puppies caused by a pathogenic variant of CCoV that was isolated from organs with severe lesions.
Figures
References
-
- Lai MMC, Holmes KV. Coronaviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields virology. 4th ed. Philadelphia: Lippincott Williams and Wilkins; 2001. p. 1163–85.
-
- Laude H, van Reeth K, Pensaert M. Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet Res. 1993;24:125–50. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
