Requirement of ELC1 for RNA polymerase II polyubiquitylation and degradation in response to DNA damage in Saccharomyces cerevisiae
- PMID: 16705154
- PMCID: PMC1489084
- DOI: 10.1128/MCB.00293-06
Requirement of ELC1 for RNA polymerase II polyubiquitylation and degradation in response to DNA damage in Saccharomyces cerevisiae
Abstract
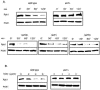
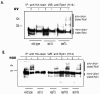
Treatment of Saccharomyces cerevisiae and human cells with DNA-damaging agents such as UV light or 4-nitroquinoline-1-oxide induces polyubiquitylation of the largest RNA polymerase II (Pol II) subunit, Rpb1, which results in rapid Pol II degradation by the proteasome. Here we identify a novel role for the yeast Elc1 protein in mediating Pol II polyubiquitylation and degradation in DNA-damaged yeast cells and propose the involvement of a ubiquitin ligase, of which Elc1 is a component, in this process. In addition, we present genetic evidence for a possible involvement of Elc1 in Rad7-Rad16-dependent nucleotide excision repair (NER) of lesions from the nontranscribed regions of the genome and suggest a role for Elc1 in increasing the proficiency of repair of nontranscribed DNA, where as a component of the Rad7-Rad16-Elc1 ubiquitin ligase, it would promote the efficient turnover of the NER ensemble from the lesion site in a Rad23-19S proteasomal complex-dependent reaction.
Figures



Similar articles
-
ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitylation and degradation in DNA-damaged yeast cells.Mol Cell Biol. 2007 Apr;27(8):3211-6. doi: 10.1128/MCB.00091-07. Epub 2007 Feb 12. Mol Cell Biol. 2007. PMID: 17296727 Free PMC article.
-
Yeast Elc1 plays an important role in global genomic repair but not in transcription coupled repair.DNA Repair (Amst). 2009 Jan 1;8(1):40-50. doi: 10.1016/j.dnarep.2008.08.010. Epub 2008 Oct 7. DNA Repair (Amst). 2009. PMID: 18817898
-
The RAD7, RAD16, and RAD23 genes of Saccharomyces cerevisiae: requirement for transcription-independent nucleotide excision repair in vitro and interactions between the gene products.Mol Cell Biol. 1997 Feb;17(2):635-43. doi: 10.1128/MCB.17.2.635. Mol Cell Biol. 1997. PMID: 9001217 Free PMC article.
-
Facilitators and Repressors of Transcription-coupled DNA Repair in Saccharomyces cerevisiae.Photochem Photobiol. 2017 Jan;93(1):259-267. doi: 10.1111/php.12655. Epub 2016 Nov 30. Photochem Photobiol. 2017. PMID: 27796045 Review.
-
A role for Rad23 proteins in 26S proteasome-dependent protein degradation?Mutat Res. 2002 Jan 29;499(1):53-61. doi: 10.1016/s0027-5107(01)00291-3. Mutat Res. 2002. PMID: 11804604 Review.
Cited by
-
A role for the Cockayne Syndrome B (CSB)-Elongin ubiquitin ligase complex in signal-dependent RNA polymerase II transcription.J Biol Chem. 2021 Jul;297(1):100862. doi: 10.1016/j.jbc.2021.100862. Epub 2021 Jun 9. J Biol Chem. 2021. PMID: 34116057 Free PMC article.
-
Nucleotide Excision Repair: Finely Tuned Molecular Orchestra of Early Pre-incision Events.Photochem Photobiol. 2017 Jan;93(1):166-177. doi: 10.1111/php.12647. Epub 2016 Nov 17. Photochem Photobiol. 2017. PMID: 27696486 Free PMC article. Review.
-
Structure of the transcribing RNA polymerase II-Elongin complex.Nat Struct Mol Biol. 2023 Dec;30(12):1925-1935. doi: 10.1038/s41594-023-01138-w. Epub 2023 Nov 6. Nat Struct Mol Biol. 2023. PMID: 37932450 Free PMC article.
-
Yeast Rpn4 Links the Proteasome and DNA Repair via RAD52 Regulation.Int J Mol Sci. 2020 Oct 30;21(21):8097. doi: 10.3390/ijms21218097. Int J Mol Sci. 2020. PMID: 33143019 Free PMC article.
-
The ubiquitin-proteasome system of Saccharomyces cerevisiae.Genetics. 2012 Oct;192(2):319-60. doi: 10.1534/genetics.112.140467. Genetics. 2012. PMID: 23028185 Free PMC article. Review.
References
-
- Aso, T., and M. N. Conrad. 1997. Molecular cloning of DNAs encoding the regulatory subunits of elongin from Saccahromyces cerevisiae and Drosophila melanogaster. Biochem. Biophys. Res. Commun. 241:334-340. - PubMed
-
- Aso, T., W. S. Lane, J. W. Conaway, and R. C. Conaway. 1995. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science 269:1439-1443. - PubMed
-
- Bradsher, J. N., K. W. Jackson, R. C. Conaway, and J. W. Conaway. 1993. RNA polymerase II transcription factor SIII. I. Identification, purification, and properties. J. Biol. Chem. 268:25587-25593. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
