SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment
- PMID: 16705163
- PMCID: PMC1489090
- DOI: 10.1128/MCB.01849-05
SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment
Abstract
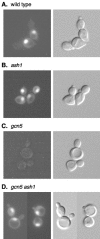
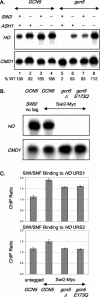
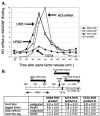
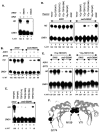
We use chromatin immunoprecipitation assays to show that the Gcn5 histone acetyltransferase in SAGA is required for SWI/SNF association with the HO promoter and that binding of SWI/SNF and SAGA are interdependent. Previous results showed that SWI/SNF binding to HO was Gcn5 independent, but that work used a strain with a mutation in the Ash1 daughter-specific repressor of HO expression. Here, we show that Ash1 functions as a repressor that inhibits SWI/SNF binding and that Gcn5 is required to overcome Ash1 repression in mother cells to allow HO transcription. Thus, Gcn5 facilitates SWI/SNF binding by antagonizing Ash1. Similarly, a mutation in SIN3, like an ash1 mutation, allows both HO expression and SWI/SNF binding in the absence of Gcn5. Although Ash1 has recently been identified in a Sin3-Rpd3 complex, our genetic analysis shows that Ash1 and Sin3 have distinct functions in regulating HO. Analysis of mutant strains shows that SWI/SNF binding and HO expression are correlated and regulated by histone acetylation. The defect in HO expression caused by a mutant SWI/SNF with a Swi2(E834K) substitution can be partially suppressed by ash1 or spt3 mutation or by a gain-of-function V71E substitution in the TATA-binding protein (TBP). Spt3 inhibits TBP binding at HO, and genetic analysis suggests that Spt3 and TBP(V71E) act in the same pathway, distinct from that of Ash1. We have detected SWI/SNF binding at the HO TATA region, and our results suggest that SWI/SNF, either directly or indirectly, facilitates TBP binding at HO.
Figures



Similar articles
-
Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex.Mol Cell Biol. 2004 Sep;24(18):8312-21. doi: 10.1128/MCB.24.18.8312-8321.2004. Mol Cell Biol. 2004. PMID: 15340090 Free PMC article.
-
Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein.Mol Cell Biol. 2003 Mar;23(6):1910-21. doi: 10.1128/MCB.23.6.1910-1921.2003. Mol Cell Biol. 2003. PMID: 12612066 Free PMC article.
-
Multiple Negative Regulators Restrict Recruitment of the SWI/SNF Chromatin Remodeler to the HO Promoter in Saccharomyces cerevisiae.Genetics. 2019 Aug;212(4):1181-1204. doi: 10.1534/genetics.119.302359. Epub 2019 Jun 5. Genetics. 2019. PMID: 31167839 Free PMC article.
-
Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
-
Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander.EMBO Rep. 2004 Oct;5(10):953-7. doi: 10.1038/sj.embor.7400251. EMBO Rep. 2004. PMID: 15459746 Free PMC article. Review.
Cited by
-
Stochastic expression and epigenetic memory at the yeast HO promoter.Proc Natl Acad Sci U S A. 2013 Aug 20;110(34):14012-7. doi: 10.1073/pnas.1306113110. Epub 2013 Jul 8. Proc Natl Acad Sci U S A. 2013. PMID: 23836672 Free PMC article.
-
Proteomic characterization of the arsenic response locus in S. cerevisiae.Epigenetics. 2019 Feb;14(2):130-145. doi: 10.1080/15592294.2019.1580110. Epub 2019 Mar 1. Epigenetics. 2019. PMID: 30739529 Free PMC article.
-
Sgf29p facilitates the recruitment of TATA box binding protein but does not alter SAGA's global structural integrity in vivo.Biochemistry. 2012 Jan 17;51(2):706-14. doi: 10.1021/bi201708z. Epub 2012 Jan 6. Biochemistry. 2012. PMID: 22224423 Free PMC article.
-
Stochastic tuning of gene expression enables cellular adaptation in the absence of pre-existing regulatory circuitry.Elife. 2018 Apr 5;7:e31867. doi: 10.7554/eLife.31867. Elife. 2018. PMID: 29620524 Free PMC article.
-
The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes.J Biol Chem. 2008 Nov 28;283(48):33101-9. doi: 10.1074/jbc.M805258200. Epub 2008 Sep 30. J Biol Chem. 2008. PMID: 18826948 Free PMC article.
References
-
- Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
