Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP
- PMID: 16705181
- PMCID: PMC1489100
- DOI: 10.1128/MCB.02183-05
Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP
Abstract
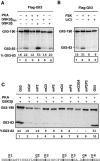
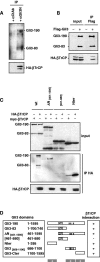
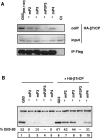
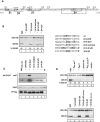
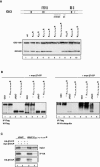
Gli3 is a zinc finger transcription factor proteolytically processed into a truncated repressor lacking C-terminal activation domains. Gli3 processing is stimulated by protein kinase A (PKA) and inhibited by Hedgehog signaling, a major signaling pathway in vertebrate development and disease. We show here that multisite glycogen synthase kinase 3beta (GSK3beta) phosphorylation and ubiquitination by SCFbetaTrCP are required for Gli3 processing. We identified multiple betaTrCP-binding sites related to the DSGX2-4S motif in Gli3, which are intertwined with PKA and GSK3beta sites, and SCFbetaTrCP target lysines that are essential for processing. Our results support a simple model whereby PKA triggers a cascade of Gli3 phosphorylation by GSK3beta and CK1 that leads to direct betaTrCP binding and ubiquitination by SCFbetaTrCP. Binding of betaTrCP to Gli3 N- and C-terminal domains lacking DSGX2-4S-related motifs was also observed, which could reflect indirect interaction via other components of Hedgehog signaling, such as the tumor suppressor Sufu. Gli3 therefore joins a small set of transcription factors whose processing is regulated by the ubiquitin-proteasome pathway. Our study sheds light on the role of PKA phosphorylation in Gli3 processing and will help to analyze how dose-dependent tuning of Gli3 processing is achieved by Hedgehog signaling.
Figures



References
-
- Bai, C. B., D. Stephen, and A. L. Joyner. 2004. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell 6:103-115. - PubMed
-
- Busino, L., M. Donzelli, M. Chiesa, D. Guardavaccaro, D. Ganoth, N. V. Dorrello, A. Hershko, M. Pagano, and G. F. Draetta. 2003. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature 426:87-91. - PubMed
-
- Chen, C. H., D. P. von Kessler, W. Park, B. Wang, Y. Ma, and P. A. Beachy. 1999. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell 98:305-316. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
