The genome of the obligately intracellular bacterium Ehrlichia canis reveals themes of complex membrane structure and immune evasion strategies
- PMID: 16707693
- PMCID: PMC1482910
- DOI: 10.1128/JB.01837-05
The genome of the obligately intracellular bacterium Ehrlichia canis reveals themes of complex membrane structure and immune evasion strategies
Abstract
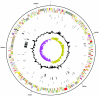
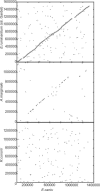
Ehrlichia canis, a small obligately intracellular, tick-transmitted, gram-negative, alpha-proteobacterium, is the primary etiologic agent of globally distributed canine monocytic ehrlichiosis. Complete genome sequencing revealed that the E. canis genome consists of a single circular chromosome of 1,315,030 bp predicted to encode 925 proteins, 40 stable RNA species, 17 putative pseudogenes, and a substantial proportion of noncoding sequence (27%). Interesting genome features include a large set of proteins with transmembrane helices and/or signal sequences and a unique serine-threonine bias associated with the potential for O glycosylation that was prominent in proteins associated with pathogen-host interactions. Furthermore, two paralogous protein families associated with immune evasion were identified, one of which contains poly(G-C) tracts, suggesting that they may play a role in phase variation and facilitation of persistent infections. Genes associated with pathogen-host interactions were identified, including a small group encoding proteins (n = 12) with tandem repeats and another group encoding proteins with eukaryote-like ankyrin domains (n = 7).
Figures

References
-
- Andersson, S. G., and C. G. Kurland. 1998. Reductive evolution of resident genomes. Trends Microbiol. 6:263-268. - PubMed
-
- Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. - PubMed
-
- Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. - PubMed
-
- Benz, I., and M. A. Schmidt. 2002. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45:267-276. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

