Shear force at the cell-matrix interface: enhanced analysis for microfabricated post array detectors
- PMID: 16708468
- PMCID: PMC1480360
Shear force at the cell-matrix interface: enhanced analysis for microfabricated post array detectors
Abstract
The interplay of mechanical forces between the extracellular environment and the cytoskeleton drives development, repair, and senescence in many tissues. Quantitative definition of these forces is a vital step in understanding cellular mechanosensing. Microfabricated post array detectors (mPADs) provide direct measurements of cell-generated forces during cell adhesion to extracellular matrix. A new approach to mPAD post labeling, volumetric imaging, and an analysis of post bending mechanics determined that cells apply shear forces and not point moments at the matrix interface. In addition, these forces could be accurately resolved from post deflections by using images of post tops and bases. Image analysis tools were then developed to increase the precision and throughput of post centroid location. These studies resulted in an improved method of force measurement with broad applicability and concise execution using a fully automated force analysis system. The new method measures cell-generated forces with less than 5% error and less than 90 seconds of computational time. Using this approach, we demonstrated direct and distinct relationships between cellular traction force and spread cell surface area for fibroblasts, endothelial cells, epithelial cells and smooth muscle cells.
Figures



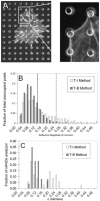

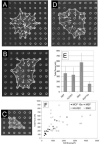
References
-
- Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3(5):466–72. - PubMed
-
- Beer F, Johnston E. Mechanics of Materials. McGraw-Hill, Inc.; 1981.
-
- Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–95. - PubMed
-
- Bogatcheva NV, Garcia JG, Verin AD. Molecular mechanisms of thrombin-induced endothelial cell permeability. Biochemistry (Mosc) 2002;67(1):75–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
