A systems approach to mapping DNA damage response pathways
- PMID: 16709784
- PMCID: PMC2811083
- DOI: 10.1126/science.1122088
A systems approach to mapping DNA damage response pathways
Abstract
Failure of cells to respond to DNA damage is a primary event associated with mutagenesis and environmental toxicity. To map the transcriptional network controlling the damage response, we measured genomewide binding locations for 30 damage-related transcription factors (TFs) after exposure of yeast to methyl-methanesulfonate (MMS). The resulting 5272 TF-target interactions revealed extensive changes in the pattern of promoter binding and identified damage-specific binding motifs. As systematic functional validation, we identified interactions for which the target changed expression in wild-type cells in response to MMS but was nonresponsive in cells lacking the TF. Validated interactions were assembled into causal pathway models that provide global hypotheses of how signaling, transcription, and phenotype are integrated after damage.
Figures
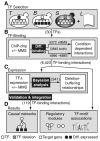

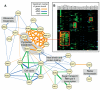


References
-
- Friedberg EC, et al. DNA Repair and Mutagenesis. ed. 2 American Society for Microbiology; Washington, DC: 2005.
-
- Rouse J, Jackson SP. Science. 2002;297:547. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

