Cigarette smoke condensate induces cytochromes P450 and aldo-keto reductases in oral cancer cells
- PMID: 16713138
- PMCID: PMC5774676
- DOI: 10.1016/j.toxlet.2006.03.008
Cigarette smoke condensate induces cytochromes P450 and aldo-keto reductases in oral cancer cells
Abstract
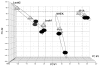
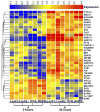
Our objective is to identify molecular factors which contribute to the increased risk of smokers for oral squamous cell carcinoma (OSCC). In the present study, we investigated the effects of cigarette smoke condensate (CSC) on gene expression profiles in different human oral cell phenotypes: normal epidermal keratinocytes (NHEK), oral dysplasia cell lines (Leuk1 and Leuk2), and a primary oral carcinoma cell line (101A). We determined differential gene expression patterns in CSC-exposed versus non-exposed cells using high-density microarray RNA expression profiling and validation by quantitative real-time RT-PCR. A set of 35 genes was specifically up- or down-regulated following CSC treatment (25microg/ml for 24h) by at least 2-fold in any one cell type. Notably, five genes of the cytochrome P450 (CYP1A1, CYP1B1) and aldo-keto reductase (AKR1C1, AKR1C3, AKR1B10) families were highly increased in expression, some of them 15- to 30-fold. The timing and extent of induction for these genes differed among the four cell phenotypes. A potential biological interaction network for the CSC response in oral cells was derived from these data, proposing novel putative response pathways. These CSC-responsive genes presumably participate in the prevention or repair of carcinogen-induced DNA damage in tobacco-related oral carcinogenesis, and may potentially be exploited for determining the severity of exposure and for correcting mutagenic damage in exposed tissues of the oral cavity.
Figures


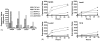

References
-
- Alexandrie AK, Sundberg MI, Seidegard J, Tornling G, Rannug A. Genetic susceptibility to lung cancer with special emphasis on CYP1A1 and GSTM1: a study on host factors in relation to age at onset, gender and histological cancer types. Carcinogenesis. 1994;15:1785–1790. - PubMed
-
- Almahmeed T, Boyle JO, Cohen EG, Carew JF, Du B, Altorki NK, Kopelovich L, Fang JL, Lazarus P, Subbaramaiah K, Dannenberg AJ. Benzo[a]pyrene phenols are more potent inducers of CYP1A1, CYP1B1 and COX-2 than benzo[a]pyrene glucuronides in cell lines derived from the human aerodigestive tract. Carcinogenesis. 2004;25:793–799. - PubMed
-
- Bartsch H, Nair U, Risch A, Rojas M, Wikman H, Alexandrov K. Genetic polymorphism of CYP genes, alone or in combination, as a risk modifier of tobacco-related cancers. Cancer Epidemiol Biomarkers Prev. 2000;9:3–28. - PubMed
-
- Burczynski ME, Sridhar GR, Palackal NT, Penning TM. The reactive oxygen species and Michael acceptor-inducible human aldo-keto reductase AKR1C1 reduces the alpha, beta-unsaturated aldehyde 4-hydroxy-2-nonenal to 1,4-dihydroxy-2-nonene. J Biol Chem. 2001;276:2890–2897. - PubMed
-
- Crofts FG, Strickland PT, Hayes CL, Sutter TR. Metabolism of 2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine (PhIP) by human cytochrome P4501B1. Carcinogenesis. 1997;18:1793–1798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

