Transcriptional pulsing of a developmental gene
- PMID: 16713960
- PMCID: PMC4764056
- DOI: 10.1016/j.cub.2006.03.092
Transcriptional pulsing of a developmental gene
Abstract
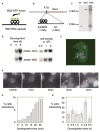
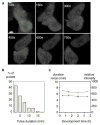
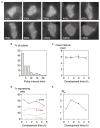
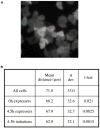
It has not been possible to view the transcriptional activity of a single gene within a living eukaryotic cell. It is therefore unclear how long and how frequently a gene is actively transcribed, how this is modulated during differentiation, and how transcriptional events are dynamically coordinated in cell populations. By means of an in vivo RNA detection technique , we have directly visualized transcription of an endogenous developmental gene. We found discrete "pulses" of gene activity that turn on and off at irregular intervals. Surprisingly, the length and height of these pulses were consistent throughout development. However, there was strong developmental variation in the proportion of cells recruited to the expressing pool. Cells were more likely to re-express than to initiate new expression, indicating that we directly observe a transcriptional memory. In addition, we used a clustering algorithm to reveal synchronous transcription initiation in neighboring cells. This study represents the first direct visualization of transcriptional pulsing in eukaryotes. Discontinuity of transcription may allow greater flexibility in the gene-expression decisions of a cell.
Figures
Comment in
-
Eukaryotic transcription: what does it mean for a gene to be 'on'?Curr Biol. 2006 May 23;16(10):R371-3. doi: 10.1016/j.cub.2006.04.014. Curr Biol. 2006. PMID: 16713947
References
-
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. - PubMed
-
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. - PubMed
-
- Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

