Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region
- PMID: 16714445
- PMCID: PMC1464109
- DOI: 10.1093/nar/gkl354
Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region
Abstract
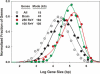
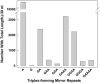
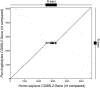
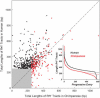
Homo(purine*pyrimidine) sequences (R*Y tracts) with mirror repeat symmetries form stable triplexes that block replication and transcription and promote genetic rearrangements. A systematic search was conducted to map the location of the longest R*Y tracts in the human genome in order to assess their potential function(s). The 814 R*Y tracts with > or =250 uninterrupted base pairs were preferentially clustered in the pseudoautosomal region of the sex chromosomes and located in the introns of 228 annotated genes whose protein products were associated with functions at the cell membrane. These genes were highly expressed in the brain and particularly in genes associated with susceptibility to mental disorders, such as schizophrenia. The set of 1957 genes harboring the 2886 R*Y tracts with > or =100 uninterrupted base pairs was additionally enriched in proteins associated with phosphorylation, signal transduction, development and morphogenesis. Comparisons of the > or =250 bp R*Y tracts in the mouse and chimpanzee genomes indicated that these sequences have mutated faster than the surrounding regions and are longer in humans than in chimpanzees. These results support a role for long R*Y tracts in promoting recombination and genome diversity during evolution through destabilization of chromosomal DNA, thereby inducing repair and mutation.
Figures


References
-
- Sinden R.R. DNA Structure and Function. San Diego, CA: Academic Press; 1994.
-
- Mirkin S.M., Frank-Kamenetskii M.D. H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 1994;23:541–576. - PubMed
-
- Rich A., Zhang S. Timeline: Z-DNA: the long road to biological function. Nature Rev. Genet. 2003;4:566–572. - PubMed
-
- Neidle S., Parkinson G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. - PubMed

