Molecular architecture of a kinetochore-microtubule attachment site
- PMID: 16715078
- PMCID: PMC2867088
- DOI: 10.1038/ncb1414
Molecular architecture of a kinetochore-microtubule attachment site
Abstract
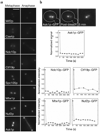
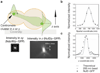
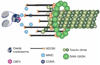
Kinetochore attachment to spindle microtubule plus-ends is necessary for accurate chromosome segregation during cell division in all eukaryotes. The centromeric DNA of each chromosome is linked to microtubule plus-ends by eight structural-protein complexes. Knowing the copy number of each of these complexes at one kinetochore-microtubule attachment site is necessary to understand the molecular architecture of the complex, and to elucidate the mechanisms underlying kinetochore function. We have counted, with molecular accuracy, the number of structural protein complexes in a single kinetochore-microtubule attachment using quantitative fluorescence microscopy of GFP-tagged kinetochore proteins in the budding yeast Saccharomyces cerevisiae. We find that relative to the two Cse4p molecules in the centromeric histone, the copy number ranges from one or two for inner kinetochore proteins such as Mif2p, to 16 for the DAM-DASH complex at the kinetochore-microtubule interface. These counts allow us to visualize the overall arrangement of a kinetochore-microtubule attachment. As most of the budding yeast kinetochore proteins have homologues in higher eukaryotes, including humans, this molecular arrangement is likely to be replicated in more complex kinetochores that have multiple microtubule attachments.
Figures

References
-
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

