Yeast Protein Interactome topology provides framework for coordinated-functionality
- PMID: 16717286
- PMCID: PMC1464412
- DOI: 10.1093/nar/gkl325
Yeast Protein Interactome topology provides framework for coordinated-functionality
Abstract
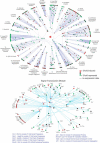
The architecture of the network of protein-protein physical interactions in Saccharomyces cerevisiae is exposed through the combination of two complementary theoretical network measures, betweenness centrality and 'Q-modularity'. The yeast interactome is characterized by well-defined topological modules connected via a small number of inter-module protein interactions. Should such topological inter-module connections turn out to constitute a form of functional coordination between the modules, we speculate that this coordination is occurring typically in a pairwise fashion, rather than by way of high-degree hub proteins responsible for coordinating multiple modules. The unique non-hub-centric hierarchical organization of the interactome is not reproduced by gene duplication-and-divergence stochastic growth models that disregard global selective pressures.
Figures



References
-
- Eisenberg D., Marcotte E.M., Xenarios I., Yeates T.O. Protein function in the post-genomic era. Nature. 2000;405:823–826. - PubMed
-
- Vidal M. A biological atlas of functional maps. Cell. 2001;104:333–339. - PubMed
-
- Bader G.D., Heilbut A., Andrews B., Tyers M., Hughes T., Boone C. Functional genomics and proteomics: charting a multidimensional map of the yeast cell. Trends Cell Biol. 2003;13:344–356. - PubMed
-
- Sharom J.R., Bellows D.S., Tyers M. From large networks to small molecules. Curr. Opin. Chem. Biol. 2004;8:81–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

