Ultrafast dynamics in DNA: "fraying" at the end of the helix
- PMID: 16719468
- PMCID: PMC2528932
- DOI: 10.1021/ja0582105
Ultrafast dynamics in DNA: "fraying" at the end of the helix
Abstract
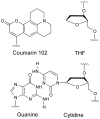
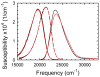
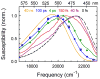
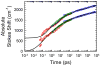
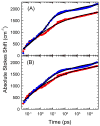
The dynamics of the electric fields in the interior of DNA are measured by using oligonucleotides in which a native base pair is replaced by a dye molecule (coumarin 102) whose emission spectrum is sensitive to the local electric field. Time-resolved measurements of the emission spectrum have been extended to a six decade time range (40 fs to 40 ns) by combining results from time-correlated photon counting, fluorescence up-conversion, and transient absorption. Recent results showed that when the reporter is placed in the center of the oligonucleotide, the dynamics are very broadly distributed over this entire time range and do not show specific time constants associated with individual processes (Andreatta, D.; et al. J. Am. Chem. Soc. 2005, 127, 7270). This paper examines an oligonucleotide with the reporter near its end. The broadly distributed relaxation seen before remains with little attenuation. In addition, a new relaxation with a well-defined relaxation time of 5 ps appears. This process is assigned to the rapid component of "fraying" at the end of the helix.
Figures




Similar articles
-
Sodium-ion binding to DNA: detection by ultrafast time-resolved stokes-shift spectroscopy.J Am Chem Soc. 2003 Oct 1;125(39):11812-3. doi: 10.1021/ja0363617. J Am Chem Soc. 2003. PMID: 14505391
-
Complex local dynamics in DNA on the picosecond and nanosecond time scales.Phys Rev Lett. 2002 Apr 15;88(15):158101. doi: 10.1103/PhysRevLett.88.158101. Epub 2002 Mar 29. Phys Rev Lett. 2002. PMID: 11955218
-
Role of monovalent counterions in the ultrafast dynamics of DNA.J Phys Chem B. 2006 Jul 6;110(26):13248-55. doi: 10.1021/jp056327+. J Phys Chem B. 2006. PMID: 16805639
-
Effect of lesions on the dynamics of DNA on the picosecond and nanosecond timescales using a polarity sensitive probe.Nucleic Acids Res. 2004 May 6;32(8):2494-507. doi: 10.1093/nar/gkh577. Print 2004. Nucleic Acids Res. 2004. PMID: 15131253 Free PMC article.
-
Toward improved biochips based on rolling circle amplification--influences of the microenvironment on the fluorescence properties of labeled DNA oligonucleotides.Ann N Y Acad Sci. 2008;1130:287-92. doi: 10.1196/annals.1430.022. Ann N Y Acad Sci. 2008. PMID: 18596361 Review.
Cited by
-
Design Considerations for RNA Spherical Nucleic Acids (SNAs).Bioconjug Chem. 2016 Sep 21;27(9):2124-31. doi: 10.1021/acs.bioconjchem.6b00350. Epub 2016 Aug 14. Bioconjug Chem. 2016. PMID: 27523252 Free PMC article.
-
modXNA: A Modular Approach to Parametrization of Modified Nucleic Acids for Use with Amber Force Fields.J Chem Theory Comput. 2024 Nov 12;20(21):9354-9363. doi: 10.1021/acs.jctc.4c01164. Epub 2024 Oct 29. J Chem Theory Comput. 2024. PMID: 39468889 Free PMC article.
-
Fluorescent analogs of biomolecular building blocks: design, properties, and applications.Chem Rev. 2010 May 12;110(5):2579-619. doi: 10.1021/cr900301e. Chem Rev. 2010. PMID: 20205430 Free PMC article. Review. No abstract available.
-
First passage time study of DNA strand displacement.Biophys J. 2021 Jun 15;120(12):2400-2412. doi: 10.1016/j.bpj.2021.01.043. Epub 2021 Apr 22. Biophys J. 2021. PMID: 33894217 Free PMC article.
-
Changes in conformational dynamics of basic side chains upon protein-DNA association.Nucleic Acids Res. 2016 Aug 19;44(14):6961-70. doi: 10.1093/nar/gkw531. Epub 2016 Jun 10. Nucleic Acids Res. 2016. PMID: 27288446 Free PMC article.
References
-
- Leroy JL, Kochoyan M, Huynhdinh T, Guéron M. J Mol Biol. 1988;200:223. - PubMed
-
- Nonin S, Leroy JL, Guéron M. Biochemistry. 1995;34:10652. - PubMed
-
- Holbrook SR, Kim SH. J Mol Biol. 1984;173:361. - PubMed
-
- Fujimoto BS, Willie ST, Reid BR, Schurr JM. J Mag Res B. 1995;106:64. - PubMed
-
- Borer PN, Laplante SR, Kumar A, Zanatta N, Martin A, Hakkinen A, Levy GC. Biochemistry. 1994;33:2441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

