Biosynthesis of isoprenoids, polyunsaturated fatty acids and flavonoids in Saccharomyces cerevisiae
- PMID: 16719921
- PMCID: PMC1533850
- DOI: 10.1186/1475-2859-5-20
Biosynthesis of isoprenoids, polyunsaturated fatty acids and flavonoids in Saccharomyces cerevisiae
Abstract
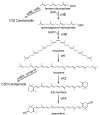
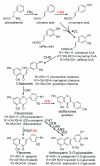
Industrial biotechnology employs the controlled use of microorganisms for the production of synthetic chemicals or simple biomass that can further be used in a diverse array of applications that span the pharmaceutical, chemical and nutraceutical industries. Recent advances in metagenomics and in the incorporation of entire biosynthetic pathways into Saccharomyces cerevisiae have greatly expanded both the fitness and the repertoire of biochemicals that can be synthesized from this popular microorganism. Further, the availability of the S. cerevisiae entire genome sequence allows the application of systems biology approaches for improving its enormous biosynthetic potential. In this review, we will describe some of the efforts on using S. cerevisiae as a cell factory for the biosynthesis of high-value natural products that belong to the families of isoprenoids, flavonoids and long chain polyunsaturated fatty acids. As natural products are increasingly becoming the center of attention of the pharmaceutical and nutraceutical industries, the use of S. cerevisiae for their production is only expected to expand in the future, further allowing the biosynthesis of novel molecular structures with unique properties.
Figures



References
-
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. - DOI - PubMed
-
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, Trochesset M, Morse D, Krogan NJ, Hiley SL, Li Z, Morris Q, Grigull J, Mitsakakis N, Roberts CJ, Greenblatt JF, Boone C, Kaiser CA, Andrews BJ, Hughes TR. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. - DOI - PubMed
-
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. - DOI - PubMed
-
- Hotze M, Schroder G, Schroder J. Cinnamate 4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P-450 proteins as translational fusions with P-450 reductase in Escherichia coli. Febs Letters. 1995;374:345–350. doi: 10.1016/0014-5793(95)01141-Z. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

