Intron gain and loss in segmentally duplicated genes in rice
- PMID: 16719932
- PMCID: PMC1779517
- DOI: 10.1186/gb-2006-7-5-r41
Intron gain and loss in segmentally duplicated genes in rice
Abstract
Background: Introns are under less selection pressure than exons, and consequently, intronic sequences have a higher rate of gain and loss than exons. In a number of plant species, a large portion of the genome has been segmentally duplicated, giving rise to a large set of duplicated genes. The recent completion of the rice genome in which segmental duplication has been documented has allowed us to investigate intron evolution within rice, a diploid monocotyledonous species.
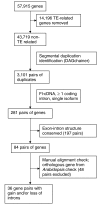
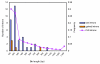
Results: Analysis of segmental duplication in rice revealed that 159 Mb of the 371 Mb genome and 21,570 of the 43,719 non-transposable element-related genes were contained within a duplicated region. In these duplicated regions, 3,101 collinear paired genes were present. Using this set of segmentally duplicated genes, we investigated intron evolution from full-length cDNA-supported non-transposable element-related gene models of rice. Using gene pairs that have an ortholog in the dicotyledonous model species Arabidopsis thaliana, we identified more intron loss (49 introns within 35 gene pairs) than intron gain (5 introns within 5 gene pairs) following segmental duplication. We were unable to demonstrate preferential intron loss at the 3' end of genes as previously reported in mammalian genomes. However, we did find that the four nucleotides of exons that flank lost introns had less frequently used 4-mers.
Conclusion: We observed that intron evolution within rice following segmental duplication is largely dominated by intron loss. In two of the five cases of intron gain within segmentally duplicated genes, the gained sequences were similar to transposable elements.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

