The TREC 2004 genomics track categorization task: classifying full text biomedical documents
- PMID: 16722582
- PMCID: PMC1440303
- DOI: 10.1186/1747-5333-1-4
The TREC 2004 genomics track categorization task: classifying full text biomedical documents
Abstract
Background: The TREC 2004 Genomics Track focused on applying information retrieval and text mining techniques to improve the use of genomic information in biomedicine. The Genomics Track consisted of two main tasks, ad hoc retrieval and document categorization. In this paper, we describe the categorization task, which focused on the classification of full-text documents, simulating the task of curators of the Mouse Genome Informatics (MGI) system and consisting of three subtasks. One subtask of the categorization task required the triage of articles likely to have experimental evidence warranting the assignment of GO terms, while the other two subtasks were concerned with the assignment of the three top-level GO categories to each paper containing evidence for these categories.
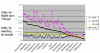
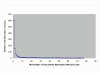
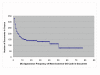
Results: The track had 33 participating groups. The mean and maximum utility measure for the triage subtask was 0.3303, with a top score of 0.6512. No system was able to substantially improve results over simply using the MeSH term Mice. Analysis of significant feature overlap between the training and test sets was found to be less than expected. Sample coverage of GO terms assigned to papers in the collection was very sparse. Determining papers containing GO term evidence will likely need to be treated as separate tasks for each concept represented in GO, and therefore require much denser sampling than was available in the data sets. The annotation subtask had a mean F-measure of 0.3824, with a top score of 0.5611. The mean F-measure for the annotation plus evidence codes subtask was 0.3676, with a top score of 0.4224. Gene name recognition was found to be of benefit for this task.
Conclusion: Automated classification of documents for GO annotation is a challenging task, as was the automated extraction of GO code hierarchies and evidence codes. However, automating these tasks would provide substantial benefit to biomedical curation, and therefore work in this area must continue. Additional experience will allow comparison and further analysis about which algorithmic features are most useful in biomedical document classification, and better understanding of the task characteristics that make automated classification feasible and useful for biomedical document curation. The TREC Genomics Track will be continuing in 2005 focusing on a wider range of triage tasks and improving results from 2004.
Figures
References
-
- Dayanik A, Fradkin D, Genkin A, Kantor P, Madigan D, Lewis DD, Menkov V. In: DIMACS at the TREC 2004 Genomics Track: ; Gaithersburg, MD. Voorhees EM and Buckland LP, editor. National Institute of Standards and Technology; 2004.
-
- Fujita S. In: Revisiting again document length hypotheses - TREC 2004 Genomics Track experiments at Patolis: ; Gaithersburg, MD. Voorhees EM and Buckland LP, editor. National Institute of Standards and Technology; 2004.
-
- Cohen AM, Bhuptiraju RT, Hersh W. In: Feature generation, feature selection, classifiers, and conceptual drift for biomedical document triage: ; Gaithersburg, MD. Voorhees EM and Buckland LP, editor. National Institute of Standards and Technology; 2004.
LinkOut - more resources
Full Text Sources
