Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer
- PMID: 16723399
- PMCID: PMC1466544
- DOI: 10.1073/pnas.0600645103
Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer
Abstract
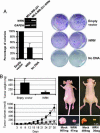
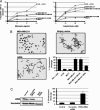
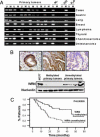
Werner syndrome (WS) is an inherited disorder characterized by premature onset of aging, genomic instability, and increased cancer incidence. The disease is caused by loss of function mutations of the WRN gene, a RecQ family member with both helicase and exonuclease activities. However, despite its putative tumor-suppressor function, little is known about the contribution of WRN to human sporadic malignancies. Here, we report that WRN function is abrogated in human cancer cells by transcriptional silencing associated with CpG island-promoter hypermethylation. We also show that, at the biochemical and cellular levels, the epigenetic inactivation of WRN leads to the loss of WRN-associated exonuclease activity and increased chromosomal instability and apoptosis induced by topoisomerase inhibitors. The described phenotype is reversed by the use of a DNA-demethylating agent or by the reintroduction of WRN into cancer cells displaying methylation-dependent silencing of WRN. Furthermore, the restoration of WRN expression induces tumor-suppressor-like features, such as reduced colony formation density and inhibition of tumor growth in nude mouse xenograft models. Screening a large collection of human primary tumors (n = 630) from different cell types revealed that WRN CpG island hypermethylation was a common event in epithelial and mesenchymal tumorigenesis. Most importantly, WRN hypermethylation in colorectal tumors was a predictor of good clinical response to the camptothecin analogue irinotecan, a topoisomerase inhibitor commonly used in the clinical setting for the treatment of this tumor type. These findings highlight the importance of WRN epigenetic inactivation in human cancer, leading to enhanced chromosomal instability and hypersensitivity to chemotherapeutic drugs.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

References
-
- Epstein C. J., Martin G. M., Schultz A. L., Motulsky A. G. Medicine (Baltimore) 1966;45:177–221. - PubMed
-
- Salk D. Hum. Genet. 1982;62:1–5. - PubMed
-
- Yu C. E., Oshima J., Fu Y. H., Wijsman E. M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., et al. Science. 1996;272:258–262. - PubMed
-
- Goto M., Imamura O., Kuromitsu J., Matsumoto T., Yamabe Y., Tokutake Y., Suzuki N., Mason B., Drayna D., Sugawara M., et al. Hum. Genet. 1997;99:191–193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

