Functional, biophysical, and structural bases for antibacterial activity of tigecycline
- PMID: 16723578
- PMCID: PMC1479133
- DOI: 10.1128/AAC.01499-05
Functional, biophysical, and structural bases for antibacterial activity of tigecycline
Abstract
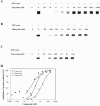
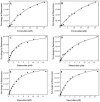
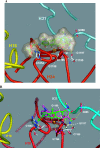
Tigecycline is a novel glycylcycline antibiotic that possesses broad-spectrum activity against many clinically relevant species of bacterial pathogens. The mechanism of action of tigecycline was delineated using functional, biophysical, and molecular modeling experiments in this study. Functional assays showed that tigecycline specifically inhibits bacterial protein synthesis with potency 3- and 20-fold greater than that of minocycline and tetracycline, respectively. Biophysical analyses demonstrated that isolated ribosomes bind tigecycline, minocycline, and tetracycline with dissociation constant values of 10(-8), 10(-7), and >10(-6) M, respectively. A molecular model of tigecycline bound to the ribosome was generated with the aid of a 3.40-angstrom resolution X-ray diffraction structure of the 30S ribosomal subunit from Thermus thermophilus. This model places tigecycline in the A site of the 30S subunit and involves substantial interactions with residues of H34 of the ribosomal subunit. These interactions were not observed in a model of tetracycline binding. Modeling data were consistent with the biochemical and biophysical data generated in this and other recent studies and suggested that tigecycline binds to bacterial ribosomes in a novel way that allows it to overcome tetracycline resistance due to ribosomal protection.
Figures




References
-
- Bauer, G., C. Berens, S. J. Projan, and W. Hillen. 2004. Comparison of tetracycline and tigecycline binding to ribosomes mapped by dimethylsulphate and drug-directed Fe2+ cleavage of 16S rRNA. J. Antimicrob. Chemother. 53:592-599. - PubMed
-
- Bergeron, J., M. Ammirati, D. Danley, L. James, M. Norcia, J. Retsema, C. A. Strick, W. G. Su, J. Sutcliffe, and L. Wondrack. 1996. Glycylcyclines bind to the high-affinity tetracycline ribosomal binding site and evade Tet(M)- and Tet(O)-mediated ribosomal protection. Antimicrob. Agents Chemother. 40:2226-2228. - PMC - PubMed
-
- Bouchillon, S. K., D. J. Hoban, B. M. Johnson, T. M. Stevens, M. J. Dowzicky, D. H. Wu, and P. A. Bradford. 2005. In vitro evaluation of tigecycline and comparative agents in 3049 clinical isolates: 2001 to 2002. Diagn. Microbiol. Infect. Dis. 51:291-295. - PubMed
-
- Bradford, P. A. 2004. Tigecycline: a first in class glycylcycline. Clin. Microbiol. Newsl. 26:163-168.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

