How initiation factors tune the rate of initiation of protein synthesis in bacteria
- PMID: 16724118
- PMCID: PMC1478179
- DOI: 10.1038/sj.emboj.7601140
How initiation factors tune the rate of initiation of protein synthesis in bacteria
Abstract
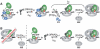
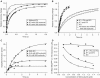
The kinetics of initiator transfer RNA (tRNA) interaction with the messenger RNA (mRNA)-programmed 30S subunit and the rate of 50S subunit docking to the 30S preinitiation complex were measured for different combinations of initiation factors in a cell-free Escherichia coli system for protein synthesis with components of high purity. The major results are summarized by a Michaelis-Menten scheme for initiation. All three initiation factors are required for maximal efficiency (kcat/KM) of initiation and for maximal in vivo rate of initiation at normal concentration of initiator tRNA. Spontaneous release of IF3 from the 30S preinitiation complex is required for subunit docking. The presence of initiator tRNA on the 30S subunit greatly increases the rate of 70S ribosome formation by increasing the rate of IF3 dissociation from the 30S subunit and the rate of 50S subunit docking to the IF3-free 30S preinitiation complex. The reasons why IF1 and IF3 are essential in E. coli are discussed in the light of the present observations.
Figures



References
-
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J (2005) The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712 - PubMed
-
- Benne R, Ebes F, Voorma HO (1973) Sequence of events in initiation of protein synthesis. Eur J Biochem 38: 265–273 - PubMed
-
- Canonaco MA, Calogero RA, Gualerzi CO (1986) Mechanism of translational initiation in prokaryotes. Evidence for a direct effect of IF2 on the activity of the 30 S ribosomal subunit. FEBS Lett 207: 198–204 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

