Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation
- PMID: 16724960
- PMCID: PMC1559456
- DOI: 10.1042/BJ20060537
Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation
Abstract
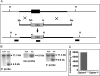
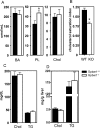
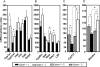
The Gpbar1 [G-protein-coupled BA (bile acid) receptor 1] is a recently identified cell-surface receptor that can bind and is activated by BAs, but its physiological role is unclear. Using targeted deletion of the Gpbar1 gene in mice, we show that the gene plays a critical role in the maintenance of bile lipid homoeostasis. Mice lacking Gpbar1 expression were viable, developed normally and did not show significant difference in the levels of cholesterol, BAs or any other bile constituents. However, they did not form cholesterol gallstones when fed a cholic acid-containing high-fat diet, and liver-specific gene expression indicated that Gpbar1-deficient mice have altered feedback regulation of BA synthesis. These results suggest that Gpbar1 plays a critical role in the formation of gallstones, possibly via a regulatory mechanism involving the cholesterol 7alpha-hydroxylase pathway.
Figures


References
-
- Chiang J. Y. Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;284:G349–G356. - PubMed
-
- Chiang J. Y. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J. Hepatol. 2004;40:539–551. - PubMed
-
- Russell D. W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003;72:137–174. - PubMed
-
- Davis R. A., Miyake J. H., Hui T. Y., Spann N. J. Regulation of cholesterol-7alpha-hydroxylase: barely missing a SHP. J. Lipid Res. 2002;43:533–543. - PubMed
-
- Fuchs M. Bile acid regulation of hepatic physiology: III. Regulation of bile acid synthesis: past progress and future challenges. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;284:G551–G557. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

