Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis
- PMID: 16725056
- PMCID: PMC1508146
- DOI: 10.1186/1471-2180-6-46
Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis
Abstract
Background: The Gram-negative bacterium Burkholderia pseudomallei (Bp) is the causative agent of the human disease melioidosis. To understand the evolutionary mechanisms contributing to Bp virulence, we performed a comparative genomic analysis of Bp K96243 and B. thailandensis (Bt) E264, a closely related but avirulent relative.
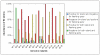
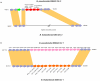
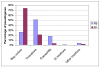
Results: We found the Bp and Bt genomes to be broadly similar, comprising two highly syntenic chromosomes with comparable numbers of coding regions (CDs), protein family distributions, and horizontally acquired genomic islands, which we experimentally validated to be differentially present in multiple Bt isolates. By examining species-specific genomic regions, we derived molecular explanations for previously-known metabolic differences, discovered potentially new ones, and found that the acquisition of a capsular polysaccharide gene cluster in Bp, a key virulence component, is likely to have occurred non-randomly via replacement of an ancestral polysaccharide cluster. Virulence related genes, in particular members of the Type III secretion needle complex, were collectively more divergent between Bp and Bt compared to the rest of the genome, possibly contributing towards the ability of Bp to infect mammalian hosts. An analysis of pseudogenes between the two species revealed that protein inactivation events were significantly biased towards membrane-associated proteins in Bt and transcription factors in Bp.
Conclusion: Our results suggest that a limited number of horizontal-acquisition events, coupled with the fine-scale functional modulation of existing proteins, are likely to be the major drivers underlying Bp virulence. The extensive genomic similarity between Bp and Bt suggests that, in some cases, Bt could be used as a possible model system for studying certain aspects of Bp behavior.
Figures




References
-
- Baar C, Eppinger M, Raddatz G, Simon J, Lanz C, Klimmek O, Nandakumar R, Gross R, Rosinus A, Keller H, Jagtap P, Linke B, Meyer F, Lederer H, Schuster SC. Complete genome sequence and analysis of Wolinella succinogenes. Proc Natl Acad Sci. 2003;100:11690–11695. doi: 10.1073/pnas.1932838100. - DOI - PMC - PubMed
-
- Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, Brubaker RR, Fowler J, Hinnebusch J, Marceau M, Medigue C, Simonet M, Chenal-Francisque V, Souza B, Dacheux D, Elliott JM, Derbise A, Hauser LJ, Garcia E. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

