Subunit architecture of multimeric complexes isolated directly from cells
- PMID: 16729021
- PMCID: PMC1479597
- DOI: 10.1038/sj.embor.7400702
Subunit architecture of multimeric complexes isolated directly from cells
Abstract
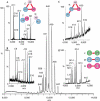
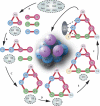
Recent developments in purification strategies, together with mass spectrometry (MS)-based proteomics, have identified numerous in vivo protein complexes and suggest the existence of many others. Standard proteomics techniques are, however, unable to describe the overall stoichiometry, subunit interactions and organization of these assemblies, because many are heterogeneous, are present at relatively low cellular abundance and are frequently difficult to isolate. We combine two existing methodologies to tackle these challenges: tandem affinity purification to isolate sufficient quantities of highly pure native complexes, and MS of the intact assemblies and subcomplexes to determine their structural organization. We optimized our protocol with two protein assemblies from Saccharomyces cerevisiae (scavenger decapping and nuclear cap-binding complexes), establishing subunit stoichiometry and identifying substoichiometric binding. We then targeted the yeast exosome, a nuclease with ten different subunits, and found that by generating subcomplexes, a three-dimensional interaction map could be derived, demonstrating the utility of our approach for large, heterogeneous cellular complexes.
Figures


References
-
- Buttner K, Wenig K, Hopfner KP (2005) Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell 20: 461–471 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

