A global view of pleiotropy and phenotypically derived gene function in yeast
- PMID: 16729036
- PMCID: PMC1681449
- DOI: 10.1038/msb4100004
A global view of pleiotropy and phenotypically derived gene function in yeast
Abstract
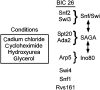
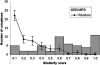
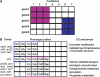
Pleiotropy, the ability of a single mutant gene to cause multiple mutant phenotypes, is a relatively common but poorly understood phenomenon in biology. Perhaps the greatest challenge in the analysis of pleiotropic genes is determining whether phenotypes associated with a mutation result from the loss of a single function or of multiple functions encoded by the same gene. Here we estimate the degree of pleiotropy in yeast by measuring the phenotypes of 4710 mutants under 21 environmental conditions, finding that it is significantly higher than predicted by chance. We use a biclustering algorithm to group pleiotropic genes by common phenotype profiles. Comparisons of these clusters to biological process classifications, synthetic lethal interactions, and protein complex data support the hypothesis that this method can be used to genetically define cellular functions. Applying these functional classifications to pleiotropic genes, we are able to dissect phenotypes into groups associated with specific gene functions.
Figures



Comment in
-
Multifunctional genes.Mol Syst Biol. 2005;1:2005.0003. doi: 10.1038/msb4100006. Epub 2005 Mar 29. Mol Syst Biol. 2005. PMID: 16729038 Free PMC article. No abstract available.
References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 - PMC - PubMed
-
- Askwith CC, de Silva D, Kaplan J (1996) Molecular biology of iron acquisition in Saccharomyces cerevisiae . Mol Microbiol 20: 27–34 - PubMed
-
- Bianchi MM, Ngo S, Vandenbol M, Sartori G, Morlupi A, Ricci C, Stefani S, Morlino GB, Hilger F, Carignani G, Slonimski PP, Frontali L (2001) Large-scale phenotypic analysis reveals identical contributions to cell functions of known and unknown yeast genes. Yeast 18: 1397–1412 - PubMed
-
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Consortium HF, Paro R, Perrimon N (2004) Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

