Phosphotyrosine interactome of the ErbB-receptor kinase family
- PMID: 16729043
- PMCID: PMC1681463
- DOI: 10.1038/msb4100012
Phosphotyrosine interactome of the ErbB-receptor kinase family
Abstract
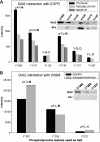
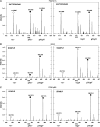
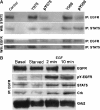
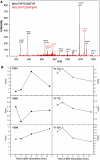
Interactions between short modified peptide motifs and modular protein domains are central events in cell signal-transduction. We determined interaction partners to all cytosolic tyrosine residues of the four members of the ErbB-receptor family in an unbiased fashion by quantitative proteomics using pull-down experiments with pairs of phosphorylated and nonphosphorylated synthetic peptides. Each receptor had characteristic preferences for interacting proteins and most interaction partners had multiple binding sites on each receptor. EGFR and ErbB4 had several docking sites for Grb2, while ErbB3 was characterized by six binding sites for PI3K. We identified STAT5 as a direct binding partner to EGFR and ErbB4 and discovered new recognition motifs for Shc and STAT5. The overall pattern of interaction partners of EGFR and ErbB4 suggests similar roles during signaling through their respective ligands. Phosphorylation kinetics of several tyrosine resides was measured by mass spectrometry and correlated with interaction partner preference. Our results demonstrate that system-wide mapping of peptide-protein interactions sites is possible, and suggest shared and unique roles of ErbB-receptor family members in downstream signaling.
Figures


Similar articles
-
Discrimination between phosphotyrosine-mediated signaling properties of conventional and neuronal Shc adapter molecules.Oncogene. 2002 Jan 3;21(1):22-31. doi: 10.1038/sj.onc.1205019. Oncogene. 2002. PMID: 11791173
-
Neuregulin-increased expression of acetylcholine receptor epsilon-subunit gene requires ErbB interaction with Shc.J Neurochem. 1999 Dec;73(6):2358-68. doi: 10.1046/j.1471-4159.1999.0732358.x. J Neurochem. 1999. PMID: 10582594
-
Mapping ErbB receptors on breast cancer cell membranes during signal transduction.J Cell Sci. 2007 Aug 15;120(Pt 16):2763-73. doi: 10.1242/jcs.007658. Epub 2007 Jul 24. J Cell Sci. 2007. PMID: 17652160
-
ErbB receptor tyrosine kinase inhibitors as therapeutic agents.Front Biosci. 2002 Sep 1;7:d1926-40. doi: 10.2741/A889. Front Biosci. 2002. PMID: 12161338 Review.
-
Receptor tyrosine kinase signaling mechanisms: Devolving TrkA responses with phosphoproteomics.Adv Biol Regul. 2013 Jan;53(1):87-96. doi: 10.1016/j.jbior.2012.10.006. Epub 2012 Nov 3. Adv Biol Regul. 2013. PMID: 23266087 Free PMC article. Review.
Cited by
-
Conditional loss of ErbB3 delays mammary gland hyperplasia induced by mutant PIK3CA without affecting mammary tumor latency, gene expression, or signaling.Cancer Res. 2013 Jul 1;73(13):4075-85. doi: 10.1158/0008-5472.CAN-12-4579. Epub 2013 Apr 30. Cancer Res. 2013. PMID: 23633485 Free PMC article.
-
Genomic Analysis of Tumors from Patients with Glioblastoma with Long-Term Response to Afatinib.Onco Targets Ther. 2022 Apr 8;15:367-380. doi: 10.2147/OTT.S346725. eCollection 2022. Onco Targets Ther. 2022. PMID: 35422631 Free PMC article.
-
Modeling cell line-specific recruitment of signaling proteins to the insulin-like growth factor 1 receptor.PLoS Comput Biol. 2019 Jan 17;15(1):e1006706. doi: 10.1371/journal.pcbi.1006706. eCollection 2019 Jan. PLoS Comput Biol. 2019. PMID: 30653502 Free PMC article.
-
Human prostatic acid phosphatase, an authentic tyrosine phosphatase, dephosphorylates ErbB-2 and regulates prostate cancer cell growth.J Biol Chem. 2010 Jul 30;285(31):23598-606. doi: 10.1074/jbc.M109.098301. Epub 2010 May 24. J Biol Chem. 2010. PMID: 20498373 Free PMC article.
-
NIMA related kinase 2 promotes gastric cancer cell proliferation via ERK/MAPK signaling.World J Gastroenterol. 2019 Jun 21;25(23):2898-2910. doi: 10.3748/wjg.v25.i23.2898. World J Gastroenterol. 2019. PMID: 31249448 Free PMC article.
References
-
- Andersen JS, Wilkinson CJ, Myoru T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 - PubMed
-
- Birge RB, Fajardo JE, Mayer BJ, Hanafusa H (1992) Tyrosine-phosphorylated epidermal growth factor receptor and cellular p130 provide high affinity binding substrates to analyze Crk-phosphotyrosine-dependent interactions in vitro. J Biol Chem 267: 10588–10595 - PubMed
-
- Blagoev B, Kratchmarova I, Ong S-E, Nielsen M, Foster L, Mann M (2003) A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol 21: 315–318 - PubMed
-
- Blagoev B, Ong S-E, Kratchmarova I, Mann M (2004) Temporal ordering of signaling networks by quantitive proteomics. Nat Biotechnol 22: 1139–1145 - PubMed
-
- Blume-Jensen P, Hunter T (2001) Oncogenic kinase signalling. Nature 411: 355–365 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

