A comprehensive pathway map of epidermal growth factor receptor signaling
- PMID: 16729045
- PMCID: PMC1681468
- DOI: 10.1038/msb4100014
A comprehensive pathway map of epidermal growth factor receptor signaling
Abstract
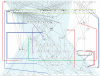
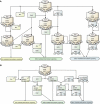
The epidermal growth factor receptor (EGFR) signaling pathway is one of the most important pathways that regulate growth, survival, proliferation, and differentiation in mammalian cells. Reflecting this importance, it is one of the best-investigated signaling systems, both experimentally and computationally, and several computational models have been developed for dynamic analysis. A map of molecular interactions of the EGFR signaling system is a valuable resource for research in this area. In this paper, we present a comprehensive pathway map of EGFR signaling and other related pathways. The map reveals that the overall architecture of the pathway is a bow-tie (or hourglass) structure with several feedback loops. The map is created using CellDesigner software that enables us to graphically represent interactions using a well-defined and consistent graphical notation, and to store it in Systems Biology Markup Language (SBML).
Figures



Comment in
-
Escalating model sizes and complexities call for standardized forms of representation.Mol Syst Biol. 2005;1:2005.0011. doi: 10.1038/msb4100015. Epub 2005 May 25. Mol Syst Biol. 2005. PMID: 16729046 Free PMC article. No abstract available.
References
-
- Carpenter G (2000) EGF receptor transactivation mediated by the proteolytic production of EGF-like agonists. Sci STKE 2000: PE1 - PubMed
-
- Chen X, Levkowitz G, Tzahar E, Karunagaran D, Lavi S, Ben-Baruch N, Leitner O, Ratzkin BJ, Bacus SS, Yarden Y (1996) An immunological approach reveals biological differences between the two NDF/heregulin receptors, ErbB-3 and ErbB-4. J Biol Chem 271: 7620–7629 - PubMed
-
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M (1999) Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol 1: 249–252 - PubMed
[References for EGFR Pathway Map] Epidermal growth factor (EGFR)
-
- Avraham H, Park SY, Schinkmann K, Avraham S (2000) RAFTK/Pyk2-mediated cellular signalling. Cell Signal 12: 123–133 - PubMed
-
- Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ (1999) c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem 274: 8335–8343 - PubMed
EGFR endocytosis followed by degradation or recycling
-
- Bao J, Alroy I, Waterman H, Schejter ED, Brodie C, Gruenberg J, Yarden Y (2000) Threonine phosphorylation diverts internalized epidermal growth factor receptors from a degradative pathway to the recycling endosome. J Biol Chem 275: 26178–26186 - PubMed
-
- Kassenbrock CK, Hunter S, Garl P, Johnson GL, Anderson SM (2002) Inhibition of Src family kinases blocks epidermal growth factor (EGF)-induced activation of Akt, phosphorylation of c-Cbl, and ubiquitination of the EGF receptor. J Biol Chem 277: 24967–24975 - PubMed
Small GTPase
-
- Ahmed S, Lee J, Kozma R, Best A, Monfries C, Lim L (1993) A novel functional target for tumor-promoting phorbol esters and lysophosphatidic acid. The p21rac-GTPase activating protein n-chimaerin. J Biol Chem 268: 10709–10712 - PubMed
-
- Burbelo PD, Drechsel D, Hall A (1995) A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem 270: 29071–29074 - PubMed
-
- Caloca MJ, Wang H, Delemos A, Wang S, Kazanietz MG (2001) Phorbol esters and related analogs regulate the subcellular localization of beta 2-chimaerin, a non-protein kinase C phorbol ester receptor. J Biol Chem 276: 18303–18312 - PubMed
-
- Cox AD, Der CJ (2003) The dark side of Ras: regulation of apoptosis. Oncogene 22: 8999–9006 - PubMed
Phosphatidylinositol phosphate (PIP) signaling
-
- Alessi DR, Deak M, Casamayor A, Caudwell FB, Morrice N, Norman DG, Gaffney P, Reese CB, MacDougall CN, Harbison D, Ashworth A, Bownes M (1997) 3-Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr Biol 7: 776–789 - PubMed
-
- Belham C, Wu S, Avruch J (1999) Intracellular signalling: PDK1–a kinase at the hub of things. Curr Biol 9: R93–R96 - PubMed
-
- Carpenter G, Ji Q (1999) Phospholipase C-gamma as a signal-transducing element. Exp Cell Res 253: 15–24 - PubMed
-
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME (1997) Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91: 231–241 - PubMed
Mitogen-activated protein kinase (MAPK) cascade
-
- Carey KD, Watson RT, Pessin JE, Stork PJ (2003) The requirement of specific membrane domains for Raf-1 phosphorylation and activation. J Biol Chem 278: 3185–3196 - PubMed
-
- Chrestensen CA, Sturgill TW (2002) Characterization of the p90 ribosomal S6 kinase 2 carboxyl-terminal domain as a protein kinase. J Biol Chem 277: 27733–27741 - PubMed
-
- Cleghon V, Morrison DK (1994) Raf-1 interacts with Fyn and Src in a non-phosphotyrosine-dependent manner. J Biol Chem 269: 17749–17755 - PubMed
-
- Crews CM, Alessandrini A, Erikson RL (1992) The primary structure of MEK, a protein kinase that phosphorylates the ERK gene product. Science 258: 478–480 - PubMed
Transcription
-
- Alvarez E, Northwood IC, Gonzalez FA, Latour DA, Seth A, Abate C, Curran T, Davis RJ (1991) Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. Characterization of the phosphorylation of c-myc and c-jun proteins by an epidermal growth factor receptor threonine 669 protein kinase. J Biol Chem 266: 15277–15285 - PubMed
-
- Coronella-Wood J, Terrand J, Sun H, Chen QM (2004) c-Fos phosphorylation induced by H2O2 prevents proteasomal degradation of c-Fos in cardiomyocytes. J Biol Chem 279: 33567–33574 - PubMed
Cell cycle
-
- Alkarain A, Jordan R, Slingerland J (2004) p27 deregulation in breast cancer: prognostic significance and implications for therapy. J Mammary Gland Biol Neoplasia 9: 67–80 - PubMed
-
- Bartkova J, Rajpert-De Meyts E, Skakkebaek NE, Lukas J, Bartek J (2003) Deregulation of the G1/S-phase control in human testicular germ cell tumours. Apmis 111: 252–265, discussion 265–256 - PubMed
-
- Bates S, Bonetta L, MacAllan D, Parry D, Holder A, Dickson C, Peters G (1994) CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene 9: 71–79 - PubMed
G protein-coupled receptor (GPCR)-mediated EGFR transactivation
-
- Beebe SJ (1994) The cAMP-dependent protein kinases and cAMP signal transduction. Semin Cancer Biol 5: 285–294 - PubMed
-
- Benzing T, Yaffe MB, Arnould T, Sellin L, Schermer B, Schilling B, Schreiber R, Kunzelmann K, Leparc GG, Kim E, Walz G (2000) 14-3-3 interacts with regulator of G protein signaling proteins and modulates their activity. J Biol Chem 275: 28167–28172 - PubMed
-
- Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA (1997) Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol 7: 191–201 - PubMed
-
- Dzimiri N (2002) Receptor crosstalk. Implications for cardiovascular function, disease and therapy. Eur J Biochem 269: 4713–4730 - PubMed
-
- Exton JH (2002) Regulation of phospholipase D. FEBS Lett 531: 58–61 - PubMed
Ca2+ signaling
-
- Banno Y, Asano T, Nozawa Y (1994) Proteolytic modification of membrane-associated phospholipase C-beta by mu-calpain enhances its activation by G-protein beta gamma subunits in human platelets. FEBS Lett 340: 185–188 - PubMed
-
- Bruce JI, Shuttleworth TJ, Giovannucci DR, Yule DI (2002) Phosphorylation of inositol 1,4,5-trisphosphate receptors in parotid acinar cells. A mechanism for the synergistic effects of cAMP on Ca2+ signaling. J Biol Chem 277: 1340–1348 - PubMed
-
- Danila CI, Hamilton SL (2004) Phosphorylation of ryanodine receptors. Biol Res 37: 521–525 - PubMed
-
- DeSouza N, Reiken S, Ondrias K, Yang YM, Matkovich S, Marks AR (2002) Protein kinase A and two phosphatases are components of the inositol 1,4,5-trisphosphate receptor macromolecular signaling complex. J Biol Chem 277: 39397–39400 - PubMed
-
- Ishida A, Shigeri Y, Taniguchi T, Kameshita I (2003) Protein phosphatases that regulate multifunctional Ca2+/calmodulin-dependent protein kinases: from biochemistry to pharmacology. Pharmacol Ther 100: 291–305 - PubMed
ErbB family
-
- Cohen BD, Green JM, Foy L, Fell HP (1996) HER4-mediated biological and biochemical properties in NIH 3T3 cells. Evidence for HER1-HER4 heterodimers. J Biol Chem 271: 4813–4818 - PubMed
-
- Elenius K, Choi CJ, Paul S, Santiestevan E, Nishi E, Klagsbrun M (1999) Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidyl inositol 3-kinase. Oncogene 18: 2607–2615 - PubMed
-
- Fiddes RJ, Campbell DH, Janes PW, Sivertsen SP, Sasaki H, Wallasch C, Daly RJ (1998) Analysis of Grb7 recruitment by heregulin-activated erbB receptors reveals a novel target selectivity for erbB3. J Biol Chem 273: 7717–7724 - PubMed
-
- Hazan R, Margolis B, Dombalagian M, Ullrich A, Zilberstein A, Schlessinger J (1990) Identification of autophosphorylation sites of HER2/neu. Cell Growth Differ 1: 3–7 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

