Refactoring bacteriophage T7
- PMID: 16729053
- PMCID: PMC1681472
- DOI: 10.1038/msb4100025
Refactoring bacteriophage T7
Abstract
Natural biological systems are selected by evolution to continue to exist and evolve. Evolution likely gives rise to complicated systems that are difficult to understand and manipulate. Here, we redesign the genome of a natural biological system, bacteriophage T7, in order to specify an engineered surrogate that, if viable, would be easier to study and extend. Our initial design goals were to physically separate and enable unique manipulation of primary genetic elements. Implicit in our design are the hypotheses that overlapping genetic elements are, in aggregate, nonessential for T7 viability and that our models for the functions encoded by elements are sufficient. To test our initial design, we replaced the left 11,515 base pairs (bp) of the 39,937 bp wild-type genome with 12,179 bp of engineered DNA. The resulting chimeric genome encodes a viable bacteriophage that appears to maintain key features of the original while being simpler to model and easier to manipulate. The viability of our initial design suggests that the genomes encoding natural biological systems can be systematically redesigned and built anew in service of scientific understanding or human intention.
Figures

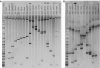

Comment in
-
Engineering novel life.Mol Syst Biol. 2005;1:2005.0020. doi: 10.1038/msb4100028. Epub 2005 Sep 13. Mol Syst Biol. 2005. PMID: 16729055 Free PMC article. No abstract available.
References
-
- Abelson H, Sussman GJ, Sussman J (1996) Structure and Interpretation of Computer Programs, 2nd edn. Cambridge, MA, USA: MIT Press
-
- Aho A-C, Donner K, Hyden C, Larsen LO, Reuter T (1988) Low retinal noise in animals with low body temperature allows high visual sensitivity. Nature 334: 348–350 - PubMed
-
- Block SM, Segall JE, Berg HC (1982) Impulse responses in bacterial chemotaxis. Cell 31: 215–226 - PubMed
-
- Carlson R (2003) The pace and proliferation of biological technologies. Biosecur Bioterror 1: 203–214 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

