PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells
- PMID: 16731587
- PMCID: PMC1488908
- DOI: 10.1105/tpc.105.035972
PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells
Erratum in
- Plant Cell. 2006 Aug;18(8):2094
Abstract
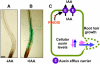
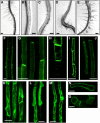
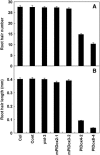
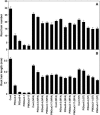
Intercellular transport of auxin is mediated by influx and efflux carriers in the plasma membrane and subjected to developmental and environmental regulation. Here, using the auxin-sensitive Arabidopsis thaliana root hair cell system and the tobacco (Nicotiana tabacum) suspension cell system, we demonstrate that the protein kinase PINOID (PID) positively regulates auxin efflux. Overexpression of PID (PIDox) or the auxin efflux carrier component PINFORMED3 (PIN3, PIN3ox), specifically in the root hair cell, greatly suppressed root hair growth. In both PIDox and PIN3ox transformants, root hair growth was nearly restored to wild-type levels by the addition of auxin, protein kinase inhibitors, or auxin efflux inhibitors. Localization of PID or PIN3 at the cell boundary was disrupted by brefeldin A and staurosporine. A mutation in the kinase domain abrogated the ability of PID to localize at the cell boundary and to inhibit root hair growth. These results suggest that PIDox- or PIN3ox-enhanced auxin efflux results in a shortage of intracellular auxin and a subsequent inhibition of root hair growth. In an auxin efflux assay using transgenic tobacco suspension cells, PIDox or PIN3ox also enhanced auxin efflux. Collectively, these results suggest that PID positively regulates cellular auxin efflux, most likely by modulating the trafficking of PIN and/or some other molecular partners involved in auxin efflux.
Figures




References
-
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11 606–612. - PubMed
-
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Arabidopsis Protocols, J.M. Martinez-Zapater and J. Salinas, eds (Totowa, NJ: Humana), pp. 259–266. - PubMed
-
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 1195–1197. - PubMed
-
- Benjamins, R., Quint, A., Weijers, D., Hooykaas, P., and Offringa, R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128 4057–4067. - PubMed
-
- Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

