tRNAHis guanylyltransferase catalyzes a 3'-5' polymerization reaction that is distinct from G-1 addition
- PMID: 16731615
- PMCID: PMC1482633
- DOI: 10.1073/pnas.0603068103
tRNAHis guanylyltransferase catalyzes a 3'-5' polymerization reaction that is distinct from G-1 addition
Abstract
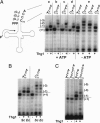
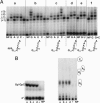
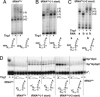
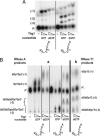
Yeast tRNA(His) guanylyltransferase, Thg1, is an essential protein that adds a single guanine to the 5' end (G(-1)) of tRNA(His). This G(-1) residue is required for aminoacylation of tRNA(His) by histidyl-tRNA synthetase, both in vitro and in vivo. The guanine nucleotide addition reaction catalyzed by Thg1 extends the polynucleotide chain in the reverse (3'-5') direction of other known polymerases, albeit by one nucleotide. Here, we show that alteration of the 3' end of the Thg1 substrate tRNA(His) unleashes an unexpected reverse polymerase activity of wild-type Thg1, resulting in the 3'-5' addition of multiple nucleotides to the tRNA, with efficiency comparable to the G(-1) addition reaction. The addition of G(-1) forms a mismatched G.A base pair at the 5' end of tRNA(His), and, with monophosphorylated tRNA substrates, it is absolutely specific for tRNA(His). By contrast, reverse polymerization forms multiple G.C or C.G base pairs, and, with preactivated tRNA species, it can initiate at positions other than -1 and is not specific for tRNA(His). Thus, wild-type Thg1 catalyzes a templated polymerization reaction acting in the reverse direction of that of canonical DNA and RNA polymerases. Surprisingly, Thg1 can also readily use dNTPs for nucleotide addition. These results suggest that 3'-5' polymerization represents either an uncharacterized role for Thg1 in RNA or DNA repair or metabolism, or it may be a remnant of an earlier catalytic strategy used in nature.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


Similar articles
-
Doing it in reverse: 3'-to-5' polymerization by the Thg1 superfamily.RNA. 2012 May;18(5):886-99. doi: 10.1261/rna.032300.112. Epub 2012 Mar 28. RNA. 2012. PMID: 22456265 Free PMC article. Review.
-
Minimal requirements for reverse polymerization and tRNA repair by tRNAHis guanylyltransferase.RNA Biol. 2018;15(4-5):614-622. doi: 10.1080/15476286.2017.1372076. Epub 2017 Sep 29. RNA Biol. 2018. PMID: 28901837 Free PMC article.
-
tRNAHis guanylyltransferase adds G-1 to the 5' end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases.RNA. 2006 Jun;12(6):1007-14. doi: 10.1261/rna.54706. Epub 2006 Apr 19. RNA. 2006. PMID: 16625026 Free PMC article.
-
The requirement for the highly conserved G-1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase.RNA. 2010 May;16(5):1068-77. doi: 10.1261/rna.2087510. Epub 2010 Apr 1. RNA. 2010. PMID: 20360392 Free PMC article.
-
The Role of 3' to 5' Reverse RNA Polymerization in tRNA Fidelity and Repair.Genes (Basel). 2019 Mar 26;10(3):250. doi: 10.3390/genes10030250. Genes (Basel). 2019. PMID: 30917604 Free PMC article. Review.
Cited by
-
Structural studies of a bacterial tRNA(HIS) guanylyltransferase (Thg1)-like protein, with nucleotide in the activation and nucleotidyl transfer sites.PLoS One. 2013 Jul 3;8(7):e67465. doi: 10.1371/journal.pone.0067465. Print 2013. PLoS One. 2013. PMID: 23844012 Free PMC article.
-
Metal ion requirement for catalysis by 3'-5' RNA polymerases.bioRxiv [Preprint]. 2025 Mar 21:2025.03.21.644660. doi: 10.1101/2025.03.21.644660. bioRxiv. 2025. Update in: Biochemistry. 2025 Aug 19;64(16):3559-3569. doi: 10.1021/acs.biochem.5c00162. PMID: 40166239 Free PMC article. Updated. Preprint.
-
tRNAHis-guanylyltransferase establishes tRNAHis identity.Nucleic Acids Res. 2012 Jan;40(1):333-44. doi: 10.1093/nar/gkr696. Epub 2011 Sep 2. Nucleic Acids Res. 2012. PMID: 21890903 Free PMC article.
-
Kinetic analysis of 3'-5' nucleotide addition catalyzed by eukaryotic tRNA(His) guanylyltransferase.Biochemistry. 2012 Jan 10;51(1):453-65. doi: 10.1021/bi201397f. Epub 2011 Dec 14. Biochemistry. 2012. PMID: 22136300 Free PMC article.
-
Doing it in reverse: 3'-to-5' polymerization by the Thg1 superfamily.RNA. 2012 May;18(5):886-99. doi: 10.1261/rna.032300.112. Epub 2012 Mar 28. RNA. 2012. PMID: 22456265 Free PMC article. Review.
References
-
- Steitz T. A. J. Biol. Chem. 1999;274:17395–17398. - PubMed
-
- Aphasizhev R., Sbicego S., Peris M., Jang S. H., Aphasizheva I., Simpson A. M., Rivlin A., Simpson L. Cell. 2002;108:637–648. - PubMed
-
- Schurer H., Schiffer S., Marchfelder A., Morl M. Biol. Chem. 2001;382:1147–1156. - PubMed
-
- Schnare M. N., Heinonen T. Y., Young P. G., Gray M. W. Curr. Genet. 1985;9:389–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

