Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription
- PMID: 16731907
- PMCID: PMC1472573
- DOI: 10.1128/JVI.02739-05
Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription
Abstract
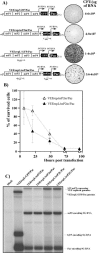
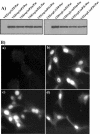
Replication of alphaviruses in vertebrate cells strongly affects cell physiology and ultimately leads to development of a cytopathic effect (CPE) and cell death. Sindbis virus (SIN) replication causes major changes in cellular macromolecular synthesis, in which the strong downregulation of transcription of cellular mRNAs and rRNAs plays a critical role. SIN nonstructural protein nsP2 was previously proposed as one of the main regulators of virus-host cell interactions, because point mutations in the carboxy-terminal part of nsP2 could make SIN and other alphaviruses and replicons less cytopathic and capable of persisting in some vertebrate cell lines. These mutants were incapable of inhibiting transcription and downregulating a viral stress-induced cell response. In the present work, we demonstrate that (i) SIN nsP2 is critically involved in CPE development, not only during the replication of SIN-specific RNAs, but also when this protein is expressed alone from different expression cassettes; (ii) the cytotoxic effect of SIN nsP2 appears to be at least partially determined by its ability to cause transcriptional shutoff; (iii) these functions of SIN nsP2 are determined by the integrity of the carboxy-terminal peptide of this protein located outside its helicase and protease domains, rather than by its protease activity; and (iv) the cytotoxic activity of SIN nsP2 depends on the presence of this protein in a free form, and alterations in P123 processing abolish the ability of nsP2 to cause CPE.
Figures





Similar articles
-
Sindbis Virus Infection Causes Cell Death by nsP2-Induced Transcriptional Shutoff or by nsP3-Dependent Translational Shutoff.J Virol. 2018 Nov 12;92(23):e01388-18. doi: 10.1128/JVI.01388-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30232189 Free PMC article.
-
Novel Mutations in nsP2 Abolish Chikungunya Virus-Induced Transcriptional Shutoff and Make the Virus Less Cytopathic without Affecting Its Replication Rates.J Virol. 2019 Feb 5;93(4):e02062-18. doi: 10.1128/JVI.02062-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30487275 Free PMC article.
-
Processing the nonstructural polyproteins of sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans.J Virol. 1989 Nov;63(11):4653-64. doi: 10.1128/JVI.63.11.4653-4664.1989. J Virol. 1989. PMID: 2529379 Free PMC article.
-
Persistent infection and suppression of host response by alphaviruses.Arch Virol Suppl. 2004;(18):139-47. doi: 10.1007/978-3-7091-0572-6_12. Arch Virol Suppl. 2004. PMID: 15119769 Review.
-
Nonstructural proteins involved in genome packaging and replication of rotaviruses and other members of the Reoviridae.Virus Res. 2004 Apr;101(1):57-66. doi: 10.1016/j.virusres.2003.12.006. Virus Res. 2004. PMID: 15010217 Review.
Cited by
-
Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component.J Virol. 2012 Mar;86(6):3121-34. doi: 10.1128/JVI.06390-11. Epub 2012 Jan 18. J Virol. 2012. PMID: 22258240 Free PMC article.
-
Self-Amplifying RNA Vaccine Candidates: Alternative Platforms for mRNA Vaccine Development.Pathogens. 2023 Jan 13;12(1):138. doi: 10.3390/pathogens12010138. Pathogens. 2023. PMID: 36678486 Free PMC article. Review.
-
Design of chimeric alphaviruses with a programmed, attenuated, cell type-restricted phenotype.J Virol. 2011 May;85(9):4363-76. doi: 10.1128/JVI.00065-11. Epub 2011 Feb 23. J Virol. 2011. PMID: 21345954 Free PMC article.
-
IRES-dependent replication of Venezuelan equine encephalitis virus makes it highly attenuated and incapable of replicating in mosquito cells.Virology. 2008 Jul 20;377(1):160-9. doi: 10.1016/j.virol.2008.04.020. Epub 2008 May 22. Virology. 2008. PMID: 18501401 Free PMC article.
-
Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning.J Virol. 2007 May;81(10):5046-57. doi: 10.1128/JVI.02746-06. Epub 2007 Feb 28. J Virol. 2007. PMID: 17329335 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

