A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus
- PMID: 16731930
- PMCID: PMC1472559
- DOI: 10.1128/JVI.00046-06
A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus
Abstract
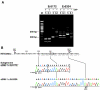
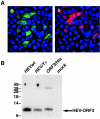
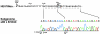
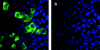
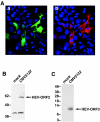
Hepatitis E virus replicons containing the neomycin resistance gene expressed from open reading frames (ORFs) 2 and 3 were transfected into Huh-7 cells, and stable cell lines containing functional replicons were selected by constant exposure to G418 sulfate. Northern blot analyses detected full-length replicon RNA and a single subgenomic RNA. This subgenomic RNA, which was capped, initiated at nucleotide 5122 downstream of the first two methionine codons in ORF3 and was bicistronic; two closely spaced methionine codons in different reading frames were used for the initiation of ORF3 and ORF2 translation.
Figures

References
-
- Agrawal, S., D. Gupta, and S. K. Panda. 2001. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp). Virology 282:87-101. - PubMed
-
- Arankalle, V. A., L. P. Chobe, M. V. Joshi, M. S. Chadha, B. Kundu, and A. M. Walimbe. 2002. Human and swine hepatitis E viruses from Western India belong to different genotypes. J. Hepatol. 36:417-425. - PubMed
-
- Balayan, M. S., A. G. Andjaparidze, S. S. Savinskaya, E. S. Ketiladze, D. M. Braginsky, A. P. Savinov, and V. F. Poleschuk. 1983. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route. Intervirology 20:23-31. - PubMed
-
- Cooper, K., F. F. Huang, L. Batista, C. D. Rayo, J. C. Bezanilla, T. E. Toth, and X. J. Meng. 2005. Identification of genotype 3 hepatitis E virus (HEV) in serum and fecal samples from pigs in Thailand and Mexico, where genotype 1 and 2 HEV strains are prevalent in the respective human populations. J. Clin. Microbiol. 43:1684-1688. - PMC - PubMed
-
- Emerson, S. U., D. Anderson, A. Arankalle, X. J. Meng, M. Purdy, G. G. Schlauder, and S. Tsarev. 2004. Hepevirus. Elsevier/Academic Press, London, United Kingdom.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

