Structure of the 21-30 fragment of amyloid beta-protein
- PMID: 16731963
- PMCID: PMC2265091
- DOI: 10.1110/ps.062076806
Structure of the 21-30 fragment of amyloid beta-protein
Abstract
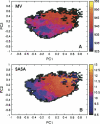
Folding and self-assembly of the 42-residue amyloid beta-protein (Abeta) are linked to Alzheimer's disease (AD). The 21-30 region of Abeta, Abeta(21-30), is resistant to proteolysis and is believed to nucleate the folding of full-length Abeta. The conformational space accessible to the Abeta(21-30) peptide is investigated by using replica exchange molecular dynamics simulations in explicit solvent. Conformations belonging to the global free energy minimum (the "native" state) from simulation are in good agreement with reported NMR structures. These conformations possess a bend motif spanning the central residues V24-K28. This bend is stabilized by a network of hydrogen bonds involving the side chain of residue D23 and the amide hydrogens of adjacent residues G25, S26, N27, and K28, as well as by a salt bridge formed between side chains of K28 and E22. The non-native states of this peptide are compact and retain a native-like bend topology. The persistence of structure in the denatured state may account for the resistance of this peptide to protease degradation and aggregation, even at elevated temperatures.
Figures

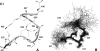

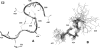

References
-
- Antzutkin O.N., Leapman R.D., Balbach J.J., Tycko R. 2002. Supramolecular structural constraints on Alzheimer's β-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance Biochemistry 41 15436–15450. - PubMed
-
- Berendsen H.J.C., van der Spoel D., van Drunen R. 1995. GROMACS: A message-passing parallel molecular dynamics implementation Comput. Phys. Commun. 91 43–56.
-
- Chimon S. and Ishii Y. 2005. Capturing intermediate structures of Alzheimer's β-amyloid, Aβ (1-40), by solid-state NMR spectroscopy J. Am. Chem. Soc. 127 13472–13473. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

