Epigenetic acquisition of inducibility of type III cytotoxicity in P. aeruginosa
- PMID: 16734902
- PMCID: PMC1488876
- DOI: 10.1186/1471-2105-7-272
Epigenetic acquisition of inducibility of type III cytotoxicity in P. aeruginosa
Abstract
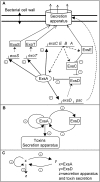
Background: Pseudomonas aeruginosa, an opportunistic pathogen, is often encountered in chronic lung diseases such as cystic fibrosis or chronic obstructive pneumonia, as well as acute settings like mechanical ventilation acquired pneumonia or neutropenic patients. It is a major cause of mortality and morbidity in these diseases. In lungs, P. aeruginosa settles in a biofilm mode of growth with the secretion of exopolysaccharides in which it is encapsulated, enhancing its antibiotic resistance and contributing to the respiratory deficiency of patients. However, bacteria must first multiply to a high density and display a cytotoxic phenotype to avoid the host's defences. A virulence determinant implicated in this step of infection is the type III secretion system (TTSS), allowing toxin injection directly into host cells. At the beginning of the infection, most strains isolated from patients' lungs possess an inducible TTSS allowing toxins injection or secretion upon in vivo or in vitro activation signals. As the infection persists most of the bacteria permanently loose this capacity, although no mutations have been evidenced. We name "non inducible" this phenotype. As suggested by the presence of a positive feedback circuit in the regulatory network controlling TTSS expression, it may be due to an epigenetic switch allowing heritable phenotypic modifications without genotype's mutations.
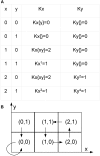
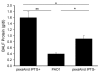
Results: Using the generalised logical method, we designed a minimal model of the TTSS regulatory network that could support the epigenetic hypothesis, and studied its dynamics which helped to define a discriminating experimental scenario sufficient to validate the epigenetic hypothesis. A mathematical framework based on formal methods from computer science allowed a rigorous validation and certification of parameters of this model leading to epigenetic behaviour. Then, we demonstrated that a non inducible strain of P. aeruginosa can stably acquire the capacity to be induced by calcium depletion for the TTSS after a short pulse of a regulatory protein. Finally, the increased cytotoxicity of a strain after this epigenetic switch was demonstrated in vivo in an acute pulmonary infection model.
Conclusion: These results may offer new perspectives for therapeutic strategies to prevent lethal infections by P. aeruginosa by reverting the epigenetic inducibility of type III cytotoxicity.
Figures
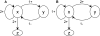

References
-
- Govan JR, Harris GS. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986;3:302–308. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

