Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors
- PMID: 16735580
- PMCID: PMC2063887
- DOI: 10.1083/jcb.200601011
Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors
Abstract
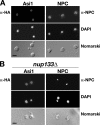
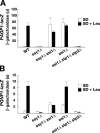
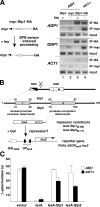
Stp1 and Stp2 are homologous transcription factors in yeast that are synthesized as latent cytoplasmic precursors with NH2-terminal regulatory domains. In response to extracellular amino acids, the plasma membrane-localized Ssy1-Ptr3-Ssy5 (SPS) sensor endoproteolytically processes Stp1 and Stp2, an event that releases the regulatory domains. The processed forms of Stp1 and Stp2 efficiently target to the nucleus and bind promoters of amino acid permease genes. In this study, we report that Asi1 is an integral component of the inner nuclear membrane that maintains the latent characteristics of unprocessed Stp1 and Stp2. In cells lacking Asi1, full-length forms of Stp1 and Stp2 constitutively induce SPS sensor-regulated genes. The regulatory domains of Stp1 and Stp2 contain a conserved motif that confers Asi1-mediated control when fused to an unrelated DNA-binding protein. Our results indicate that latent precursor forms of Stp1 and Stp2 inefficiently enter the nucleus; however, once there, Asi1 restricts them from binding SPS sensor-regulated promoters. These findings reveal an unanticipated role of inner nuclear membrane proteins in controlling gene expression.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

