A wing expressed sequence tag resource for Bicyclus anynana butterflies, an evo-devo model
- PMID: 16737530
- PMCID: PMC1534037
- DOI: 10.1186/1471-2164-7-130
A wing expressed sequence tag resource for Bicyclus anynana butterflies, an evo-devo model
Abstract
Background: Butterfly wing color patterns are a key model for integrating evolutionary developmental biology and the study of adaptive morphological evolution. Yet, despite the biological, economical and educational value of butterflies they are still relatively under-represented in terms of available genomic resources. Here, we describe an Expression Sequence Tag (EST) project for Bicyclus anynana that has identified the largest available collection to date of expressed genes for any butterfly.
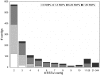
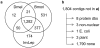
Results: By targeting cDNAs from developing wings at the stages when pattern is specified, we biased gene discovery towards genes potentially involved in pattern formation. Assembly of 9,903 ESTs from a subtracted library allowed us to identify 4,251 genes of which 2,461 were annotated based on BLAST analyses against relevant gene collections. Gene prediction software identified 2,202 peptides, of which 215 longer than 100 amino acids had no homology to any known proteins and, thus, potentially represent novel or highly diverged butterfly genes. We combined gene and Single Nucleotide Polymorphism (SNP) identification by constructing cDNA libraries from pools of outbred individuals, and by sequencing clones from the 3' end to maximize alignment depth. Alignments of multi-member contigs allowed us to identify over 14,000 putative SNPs, with 316 genes having at least one high confidence double-hit SNP. We furthermore identified 320 microsatellites in transcribed genes that can potentially be used as genetic markers.
Conclusion: Our project was designed to combine gene and sequence polymorphism discovery and has generated the largest gene collection available for any butterfly and many potential markers in expressed genes. These resources will be invaluable for exploring the potential of B. anynana in particular, and butterflies in general, as models in ecological, evolutionary, and developmental genetics.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

