Identification of multiple distinct Snf2 subfamilies with conserved structural motifs
- PMID: 16738128
- PMCID: PMC1474054
- DOI: 10.1093/nar/gkl295
Identification of multiple distinct Snf2 subfamilies with conserved structural motifs
Abstract
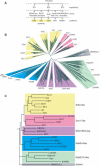
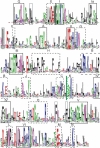
The Snf2 family of helicase-related proteins includes the catalytic subunits of ATP-dependent chromatin remodelling complexes found in all eukaryotes. These act to regulate the structure and dynamic properties of chromatin and so influence a broad range of nuclear processes. We have exploited progress in genome sequencing to assemble a comprehensive catalogue of over 1300 Snf2 family members. Multiple sequence alignment of the helicase-related regions enables 24 distinct subfamilies to be identified, a considerable expansion over earlier surveys. Where information is known, there is a good correlation between biological or biochemical function and these assignments, suggesting Snf2 family motor domains are tuned for specific tasks. Scanning of complete genomes reveals all eukaryotes contain members of multiple subfamilies, whereas they are less common and not ubiquitous in eubacteria or archaea. The large sample of Snf2 proteins enables additional distinguishing conserved sequence blocks within the helicase-like motor to be identified. The establishment of a phylogeny for Snf2 proteins provides an opportunity to make informed assignments of function, and the identification of conserved motifs provides a framework for understanding the mechanisms by which these proteins function.
Figures

References
-
- Gorbalenya A.E., Koonin E.V. Helicases: amino acid sequence comparisons and structure–function relationship. Curr. Opin. Struct. Biol. 1993;3:419–429.
-
- Tanner N.K., Cordin O., Banroques J., Doere M., Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. - PubMed
-
- Subramanya H.S., Bird L.E., Brannigan J.A., Wigley D.B. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. - PubMed

