Centromeric histone H3 is essential for vegetative cell division and for DNA elimination during conjugation in Tetrahymena thermophila
- PMID: 16738316
- PMCID: PMC1489134
- DOI: 10.1128/MCB.00079-06
Centromeric histone H3 is essential for vegetative cell division and for DNA elimination during conjugation in Tetrahymena thermophila
Abstract
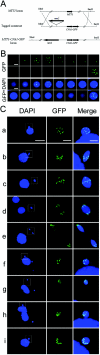
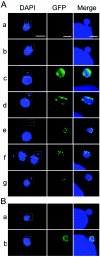
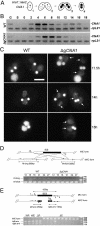
The Tetrahymena thermophila CNA1 gene encodes the centromeric H3, Cna1p. Green fluorescent protein (GFP)-tagged Cna1p localizes in micronuclei in dots whose number and behavior during mitosis and conjugation are consistent with centromeres. During interphase, Cna1p-GFP localizes in peripheral dots, suggesting centromeres are associated with the nuclear envelope. Newly synthesized Cna1p-GFP enters micronuclei in mitosis and accumulates in the nucleoplasm. Its deposition at centromeres starts at early S phase and continues through most of S phase. CNA1 is required for vegetative cell growth. Knockdown of CNA1 genes in the somatic macronucleus results in micronuclear DNA loss and delayed chromosome segregation during mitosis. During conjugation, Cna1p-GFP disappears from the centromeres in the developing macronucleus, consistent with centromeric sequences being internal eliminated sequences. Surprisingly, zygotic CNA1 is required for efficient elimination of germ line-specific sequences during development of the new macronuclei but not for the RNA interference pathway, through which sequences are targeted for elimination. Zygotically expressed Cna1p localizes in the spherical structures in which the later stages of DNA elimination occur, and these structures cannot be formed in the absence of zygotic CNA1, suggesting that, in addition to functioning in centromeres, Cna1p may also play a role in organizing the formation of the DNA elimination structures.
Figures




Similar articles
-
The CNA1 histone of the ciliate Tetrahymena thermophila is essential for chromosome segregation in the germline micronucleus.Mol Biol Cell. 2006 Jan;17(1):485-97. doi: 10.1091/mbc.e05-07-0698. Epub 2005 Oct 26. Mol Biol Cell. 2006. PMID: 16251352 Free PMC article.
-
Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila.Mol Cell Biol. 2005 Oct;25(20):9151-64. doi: 10.1128/MCB.25.20.9151-9164.2005. Mol Cell Biol. 2005. PMID: 16199890 Free PMC article.
-
Zygotic expression of the double-stranded RNA binding motif protein Drb2p is required for DNA elimination in the ciliate Tetrahymena thermophila.Eukaryot Cell. 2011 Dec;10(12):1648-59. doi: 10.1128/EC.05216-11. Epub 2011 Oct 21. Eukaryot Cell. 2011. PMID: 22021239 Free PMC article.
-
Developmental progression of Tetrahymena through the cell cycle and conjugation.Methods Cell Biol. 2012;109:177-236. doi: 10.1016/B978-0-12-385967-9.00007-4. Methods Cell Biol. 2012. PMID: 22444146 Review.
-
Small RNAs in genome rearrangement in Tetrahymena.Curr Opin Genet Dev. 2004 Apr;14(2):181-7. doi: 10.1016/j.gde.2004.01.004. Curr Opin Genet Dev. 2004. PMID: 15196465 Review.
Cited by
-
Histone variants--ancient wrap artists of the epigenome.Nat Rev Mol Cell Biol. 2010 Apr;11(4):264-75. doi: 10.1038/nrm2861. Epub 2010 Mar 3. Nat Rev Mol Cell Biol. 2010. PMID: 20197778 Review.
-
Tetrahymena meiotic nuclear reorganization is induced by a checkpoint kinase-dependent response to DNA damage.Mol Biol Cell. 2009 May;20(9):2428-37. doi: 10.1091/mbc.e08-10-1058. Epub 2009 Mar 18. Mol Biol Cell. 2009. PMID: 19297526 Free PMC article.
-
Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes.Res Microbiol. 2011 Jul-Aug;162(6):578-86. doi: 10.1016/j.resmic.2011.05.001. Epub 2011 May 18. Res Microbiol. 2011. PMID: 21624459 Free PMC article. Review.
-
Whats, hows and whys of programmed DNA elimination in Tetrahymena.Open Biol. 2017 Oct;7(10):170172. doi: 10.1098/rsob.170172. Open Biol. 2017. PMID: 29021213 Free PMC article. Review.
-
A germline-limited piggyBac transposase gene is required for precise excision in Tetrahymena genome rearrangement.Nucleic Acids Res. 2017 Sep 19;45(16):9481-9502. doi: 10.1093/nar/gkx652. Nucleic Acids Res. 2017. PMID: 28934495 Free PMC article.
References
-
- Black, B. E., D. R. Foltz, S. Chakravarthy, K. Luger, V. L. Woods, Jr., and D. W. Cleveland. 2004. Structural determinants for generating centromeric chromatin. Nature 430:578-582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
