Residues crucial for maintaining short paths in network communication mediate signaling in proteins
- PMID: 16738564
- PMCID: PMC1681495
- DOI: 10.1038/msb4100063
Residues crucial for maintaining short paths in network communication mediate signaling in proteins
Abstract
Here, we represent protein structures as residue interacting networks, which are assumed to involve a permanent flow of information between amino acids. By removal of nodes from the protein network, we identify fold centrally conserved residues, which are crucial for sustaining the shortest pathways and thus play key roles in long-range interactions. Analysis of seven protein families (myoglobins, G-protein-coupled receptors, the trypsin class of serine proteases, hemoglobins, oligosaccharide phosphorylases, nuclear receptor ligand-binding domains and retroviral proteases) confirms that experimentally many of these residues are important for allosteric communication. The agreement between the centrally conserved residues, which are key in preserving short path lengths, and residues experimentally suggested to mediate signaling further illustrates that topology plays an important role in network communication. Protein folds have evolved under constraints imposed by function. To maintain function, protein structures need to be robust to mutational events. On the other hand, robustness is accompanied by an extreme sensitivity at some crucial sites. Thus, here we propose that centrally conserved residues, whose removal increases the characteristic path length in protein networks, may relate to the system fragility.
Figures



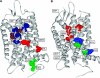


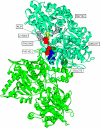


References
-
- Achacoso TB, Yamamoto WS (1992) AY's Neuroanatomy of C. elegans for Computation. Boca Raton, FL: CRC Press
-
- Aharoni A, Gaidukov L, Khersonsky O, Goulds MMcQ, Roodveldt C, Tawfik DS (2005) The evolvability of promiscuous protein functions. Nat Genet 37: 73–76 - PubMed
-
- Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S (2004) Network analysis of protein structures identifies functional residues. J Mol Biol 344: 1135–1146 - PubMed
-
- Andres A, Kosoy A, Garriga P, Manyosa J (2001) Mutations at position 125 in transmembrane helix III of rhodopsin affect the structure and signalling of the receptor. Eur J Biochem 268: 5696–5704 - PubMed
-
- Ballesteros JA, Shi L, Javitch JA (2001) Structural mimicry in G protein-coupled receptors: implications of the high-resolution structure of rhodopsin for structure–function analysis of rhodopsin-like receptors. Mol Pharmacol 60: 1–19 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

