Proteome analysis of yeast response to various nutrient limitations
- PMID: 16738570
- PMCID: PMC1681501
- DOI: 10.1038/msb4100069
Proteome analysis of yeast response to various nutrient limitations
Abstract
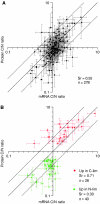
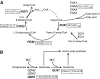
We compared the response of Saccharomyces cerevisiae to carbon (glucose) and nitrogen (ammonia) limitation in chemostat cultivation at the proteome level. Protein levels were differentially quantified using unlabeled and 15N metabolically labeled yeast cultures. A total of 928 proteins covering a wide range of isoelectric points, molecular weights and subcellular localizations were identified. Stringent statistical analysis identified 51 proteins upregulated in response to glucose limitation and 51 upregulated in response to ammonia limitation. Under glucose limitation, typical glucose-repressed genes encoding proteins involved in alternative carbon source utilization, fatty acids beta-oxidation and oxidative phosphorylation displayed an increased protein level. Proteins upregulated in response to nitrogen limitation were mostly involved in scavenging of alternative nitrogen sources and protein degradation. Comparison of transcript and protein levels clearly showed that upregulation in response to glucose limitation was mainly transcriptionally controlled, whereas upregulation in response to nitrogen limitation was essentially controlled at the post-transcriptional level by increased translational efficiency and/or decreased protein degradation. These observations underline the need for multilevel analysis in yeast systems biology.
Figures
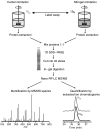

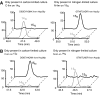
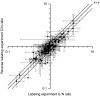
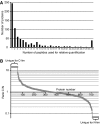
References
-
- Berger SJ, Lee SW, Anderson GA, Pasa-Tolic L, Tolic N, Shen Y, Zhao R, Smith RD (2002) High-throughput global peptide proteomic analysis by combining stable isotope amino acid labeling and data-dependent multiplexed-MS/MS. Anal Chem 74: 4994–5000 - PubMed
-
- Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M (2003) A proteomics strategy to elucidate functional protein–protein interactions applied to EGF signaling. Nat Biotechnol 21: 315–318 - PubMed
-
- Boer VM, de Winde JH, Pronk JT, Piper MD (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem 278: 3265–3274 - PubMed
-
- Conrads TP, Alving K, Veenstra TD, Belov ME, Anderson GA, Anderson DJ, Lipton MS, Pasa-Tolic L, Udseth HR, Chrisler WB, Thrall BD, Smith RD (2001) Quantitative analysis of bacterial and mammalian proteomes using a combination of cysteine affinity tags and 15N-metabolic labeling. Anal Chem 73: 2132–2139 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

