Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis
- PMID: 16740930
- PMCID: PMC1482941
- DOI: 10.1128/JB.01660-05
Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis
Abstract
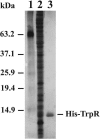
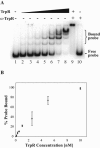
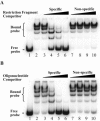
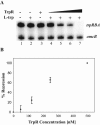
Tryptophan is an essential amino acid that is required for normal development in Chlamydia species, and tryptophan metabolism has been implicated in chlamydial persistence and tissue tropism. The ability to synthesize tryptophan is not universal among the Chlamydiaceae, but species that have a predicted tryptophan biosynthetic pathway also encode an ortholog of TrpR, a regulator of tryptophan metabolism in many gram-negative bacteria. We show that in Chlamydia trachomatis serovar D, TrpR regulates its own gene and trpB and trpA, the genes for the two subunits of tryptophan synthase. These three genes form an operon that is transcribed by the major form of chlamydial RNA polymerase. TrpR acts as a tryptophan-dependent aporepressor that binds specifically to operator sequences upstream of the trpRBA operon. We also found that TrpR repressed in vitro transcription of trpRBA in a promoter-specific manner, and the level of repression was dependent upon the concentrations of TrpR and tryptophan. Our findings provide a mechanism for chlamydiae to sense changes in tryptophan levels and to respond by modulating expression of the tryptophan biosynthesis genes, and we present a unified model that shows how C. trachomatis can combine transcriptional repression and attenuation to regulate intrachlamydial tryptophan levels. In the face of host defense mechanisms that limit tryptophan availability from the infected cell, the ability to maintain homeostatic control of intrachlamydial tryptophan levels is likely to play an important role in chlamydial pathogenesis.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

