Effect of repeat copy number on variable-number tandem repeat mutations in Escherichia coli O157:H7
- PMID: 16740932
- PMCID: PMC1482962
- DOI: 10.1128/JB.00001-06
Effect of repeat copy number on variable-number tandem repeat mutations in Escherichia coli O157:H7
Abstract
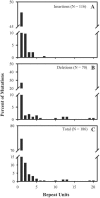
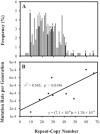
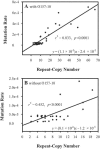
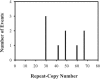
Variable-number tandem repeat (VNTR) loci have shown a remarkable ability to discriminate among isolates of the recently emerged clonal pathogen Escherichia coli O157:H7, making them a very useful molecular epidemiological tool. However, little is known about the rates at which these sequences mutate, the factors that affect mutation rates, or the mechanisms by which mutations occur at these loci. Here, we measure mutation rates for 28 VNTR loci and investigate the effects of repeat copy number and mismatch repair on mutation rate using in vitro-generated populations for 10 E. coli O157:H7 strains. We find single-locus rates as high as 7.0 x 10(-4) mutations/generation and a combined 28-locus rate of 6.4 x 10(-4) mutations/generation. We observed single- and multirepeat mutations that were consistent with a slipped-strand mispairing mutation model, as well as a smaller number of large repeat copy number mutations that were consistent with recombination-mediated events. Repeat copy number within an array was strongly correlated with mutation rate both at the most mutable locus, O157-10 (r2= 0.565, P = 0.0196), and across all mutating loci. The combined locus model was significant whether locus O157-10 was included (r2= 0.833, P < 0.0001) or excluded (r2= 0.452, P < 0.0001) from the analysis. Deficient mismatch repair did not affect mutation rate at any of the 28 VNTRs with repeat unit sizes of >5 bp, although a poly(G) homomeric tract was destabilized in the mutS strain. Finally, we describe a general model for VNTR mutations that encompasses insertions and deletions, single- and multiple-repeat mutations, and their relative frequencies based upon our empirical mutation rate data.
Figures

References
-
- Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 101:17837-17842. - PMC - PubMed
-
- Eisen, J. 1999. Mechanistic basis for microsatellite instability, p. 34-48. In D. B. Goldstein and C. Schlötterer (ed.), Microsatellites: evolution and applications. Oxford University Press, New York, N.Y.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

