RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis
- PMID: 16741558
- PMCID: PMC1472700
- DOI: 10.1371/journal.pgen.0020083
RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis
Abstract
Cytosine DNA methylation protects eukaryotic genomes by silencing transposons and harmful DNAs, but also regulates gene expression during normal development. Loss of CG methylation in the Arabidopsis thaliana met1 and ddm1 mutants causes varied and stochastic developmental defects that are often inherited independently of the original met1 or ddm1 mutation. Loss of non-CG methylation in plants with combined mutations in the DRM and CMT3 genes also causes a suite of developmental defects. We show here that the pleiotropic developmental defects of drm1 drm2 cmt3 triple mutant plants are fully recessive, and unlike phenotypes caused by met1 and ddm1, are not inherited independently of the drm and cmt3 mutations. Developmental phenotypes are also reversed when drm1 drm2 cmt3 plants are transformed with DRM2 or CMT3, implying that non-CG DNA methylation is efficiently re-established by sequence-specific signals. We provide evidence that these signals include RNA silencing though the 24-nucleotide short interfering RNA (siRNA) pathway as well as histone H3K9 methylation, both of which converge on the putative chromatin-remodeling protein DRD1. These signals act in at least three partially intersecting pathways that control the locus-specific patterning of non-CG methylation by the DRM2 and CMT3 methyltransferases. Our results suggest that non-CG DNA methylation that is inherited via a network of persistent targeting signals has been co-opted to regulate developmentally important genes.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
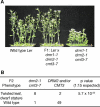





References
-
- Kakutani T. Genetic characterization of late-flowering traits induced by DNA hypomethylation mutation in Arabidopsis thaliana . Plant J. 1997;12:1447–1451. - PubMed
-
- Saze H, Scheid OM, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34:65–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

