Conformational suppression of inter-receptor signaling defects
- PMID: 16751275
- PMCID: PMC1482603
- DOI: 10.1073/pnas.0602135103
Conformational suppression of inter-receptor signaling defects
Abstract
Motile bacteria follow gradients of attractant and repellent chemicals with high sensitivity. Their chemoreceptors are physically clustered, which may enable them to function as a cooperative array. Although native chemoreceptor molecules are typically transmembrane homodimers, they appear to associate through their cytoplasmic tips to form trimers of dimers, which may be an important architectural element in the assembly and operation of receptor clusters. The five receptors of Escherichia coli that mediate most of its chemotactic and aerotactic behaviors have identical trimer contact residues and have been shown by in vivo crosslinking methods to form mixed trimers of dimers. Mutations at the trimer contact sites of Tsr, the serine chemoreceptor, invariably abrogate Tsr function, but some of those lesions (designated Tsr*) are epistatic and block the function of heterologous chemoreceptors. We isolated and characterized mutations (designated Tar()) in the aspartate chemoreceptor that restored function to Tsr* receptors. The suppressors arose at or near the Tar trimer contact sites and acted in an allele-specific fashion on Tsr* partners. Alone, many Tar() receptors were unable to mediate chemotactic responses to aspartate, but all formed clusters with varying efficiencies. Most of those Tar() receptors were epistatic to WT Tsr, but some regained Tar function in combination with a suppressible Tsr* partner. Tar()-Tsr* suppression most likely occurs through compensatory changes in the conformation or dynamics of a mixed receptor signaling complex, presumably based on trimer-of-dimer interactions. These collaborative teams may be responsible for the high-gain signaling properties of bacterial chemoreceptors.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


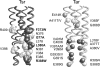

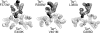
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

