The PKS4 gene of Fusarium graminearum is essential for zearalenone production
- PMID: 16751498
- PMCID: PMC1489647
- DOI: 10.1128/AEM.00963-05
The PKS4 gene of Fusarium graminearum is essential for zearalenone production
Abstract
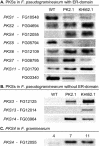
Zearalenones are produced by several Fusarium species and can cause reproductive problems in animals. Some aurofusarin mutants of Fusarium pseudograminearum produce elevated levels of zearalenone (ZON), one of the estrogenic mycotoxins comprising the zearalenones. An analysis of transcripts from polyketide synthase genes identified in the Fusarium graminearum database was carried out for these mutants. PKS4 was the only gene with an enoyl reductase domain that had a higher level of transcription in the aurofusarin mutants than in the wild type. An Agrobacterium tumefaciens-mediated transformation protocol was used to replace the central part of the PKS4 gene with a hygB resistance gene through double homologous recombination in an F. graminearum strain producing a high level of ZON. PCR and Southern analysis of transformants were used to identify isolates with single insertional replacements of PKS4. High-performance liquid chromatography analysis showed that the PKS4 replacement mutant did not produce ZON. Thus, PKS4 encodes an enzyme required for the production of ZON in F. graminearum. Barley root infection studies revealed no alteration in the pathogenicity of the PKS4 mutant compared to the pathogenicity of the wild type. The expression of PKS13, which is located in the same cluster as PKS4, decreased dramatically in the mutant, while transcription of PKS4 was unchanged. This differential expression may indicate that ZON or its derivatives do not regulate expression of PKS4 and that the PKS4-encoded protein or its product stimulates expression of PKS13. Furthermore, both the lack of aurofusarin and ZON influenced the expression of other polyketide synthases, demonstrating that one polyketide can influence the expression of others.
Figures




Similar articles
-
Real-time quantitative expression studies of the zearalenone biosynthetic gene cluster in Fusarium graminearum.Phytopathology. 2009 Feb;99(2):176-84. doi: 10.1094/PHYTO-99-2-0176. Phytopathology. 2009. PMID: 19159310
-
Intra-strains diversity of expression of polymorphic PKS4 gene in comparison in zearalenone production by Fusarium graminearum during in vitro cultivation.Acta Biochim Pol. 2016;63(1):97-102. doi: 10.18388/abp.2015_1022. Epub 2015 Oct 23. Acta Biochim Pol. 2016. PMID: 26495440
-
Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex.Fungal Genet Biol. 2005 May;42(5):420-33. doi: 10.1016/j.fgb.2005.01.010. Fungal Genet Biol. 2005. PMID: 15809006
-
Autoregulation of ZEB2 expression for zearalenone production in Fusarium graminearum.Mol Microbiol. 2015 Sep;97(5):942-56. doi: 10.1111/mmi.13078. Epub 2015 Jun 25. Mol Microbiol. 2015. PMID: 26036360
-
Quantification of Fusarium graminearum and Fusarium culmorum by real-time PCR system and zearalenone assessment in maize.Int J Food Microbiol. 2012 Mar 1;154(1-2):59-65. doi: 10.1016/j.ijfoodmicro.2011.12.022. Epub 2011 Dec 21. Int J Food Microbiol. 2012. PMID: 22240058
Cited by
-
Nonribosomal peptide synthetase (NPS) genes in Fusarium graminearum, F. culmorum and F. pseudograminearium and identification of NPS2 as the producer of ferricrocin.Curr Genet. 2007 Jan;51(1):43-58. doi: 10.1007/s00294-006-0103-0. Epub 2006 Oct 17. Curr Genet. 2007. PMID: 17043871
-
Studying the Ability of Thymol to Improve Fungicidal Effects of Tebuconazole and Difenoconazole Against Some Plant Pathogenic Fungi in Seed or Foliar Treatments.Front Microbiol. 2021 Feb 25;12:629429. doi: 10.3389/fmicb.2021.629429. eCollection 2021. Front Microbiol. 2021. PMID: 33717020 Free PMC article.
-
Fusarium culmorum: causal agent of foot and root rot and head blight on wheat.Mol Plant Pathol. 2013 May;14(4):323-41. doi: 10.1111/mpp.12011. Epub 2012 Dec 28. Mol Plant Pathol. 2013. PMID: 23279114 Free PMC article. Review.
-
Comprehensive Description of Fusarium graminearum Pigments and Related Compounds.Foods. 2018 Oct 5;7(10):165. doi: 10.3390/foods7100165. Foods. 2018. PMID: 30301164 Free PMC article. Review.
-
Secretomics identifies Fusarium graminearum proteins involved in the interaction with barley and wheat.Mol Plant Pathol. 2012 Jun;13(5):445-53. doi: 10.1111/j.1364-3703.2011.00759.x. Epub 2011 Nov 1. Mol Plant Pathol. 2012. PMID: 22044785 Free PMC article.
References
-
- Ausubel, F. M., R. E. Kingston, D. D. Moore, J. G. Seidman, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons Inc., Hoboken, N.J.
-
- Bell, A. A., M. H. Wheeler, J. G. Liu, and R. D. Stipanovic. 2003. United States Department of Agriculture Agricultural Research Service studies on polyketide toxins of Fusarium oxysporum f. sp. vasinfectum: potential targets for disease control. Pest. Manag. Sci. 59:736-747. - PubMed
-
- Bhatnagar, D., K. C. Ehrlich, and T. E. Cleveland. 2003. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 61:83-93. - PubMed
-
- Bingle, L. E. H., T. J. Simpson, and C. M. Lazarus. 1999. Ketosynthase domain probes identify two subclasses of fungal polyketide synthase genes. Fungal Genet. Biol. 26:209-223. - PubMed
-
- Blaney, B. J., and R. L. Dodman. 2002. Production of zearalenone, deoxynivalenol, nivalenol, and acetylated derivatives by Australian isolates of Fusarium graminearum and F. pseudograminearum in relation to source and culturing conditions. Aust. J. Agric. Res. 53:1317-1326.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

