Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves
- PMID: 16751500
- PMCID: PMC1489609
- DOI: 10.1128/AEM.02770-05
Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves
Abstract
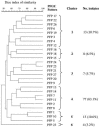
Healthy calves (n = 96, 1 to 9 weeks old) from a dairy herd in central Pennsylvania were examined each month over a five-month period for fecal shedding of ceftiofur-resistant gram-negative bacteria. Ceftiofur-resistant Escherichia coli isolates (n = 122) were characterized by antimicrobial resistance (disk diffusion and MIC), serotype, pulsed-field gel electrophoresis subtypes, beta-lactamase genes, and virulence genes. Antibiotic disk diffusion assays showed that the isolates were resistant to ampicillin (100%), ceftiofur (100%), chloramphenicol (94%), florfenicol (93%), gentamicin (89%), spectinomycin (72%), tetracycline (98%), ticarcillin (99%), and ticarcillin-clavulanic acid (99%). All isolates were multidrug resistant and displayed elevated MICs. The E. coli isolates belonged to 42 serotypes, of which O8:H25 was the predominant serotype (49.2%). Pulsed-field gel electrophoresis classified the E. coli isolates into 27 profiles. Cluster analysis showed that 77 isolates (63.1%) belonged to one unique group. The prevalence of pathogenic E. coli was low (8%). A total of 117 ceftiofur-resistant E. coli isolates (96%) possessed the bla(CMY2) gene. Based on phenotypic and genotypic characterization, the ceftiofur-resistant E. coli isolates belonged to 59 clonal types. There was no significant relationship between calf age and clonal type. The findings of this study revealed that healthy dairy calves were rapidly colonized by antibiotic-resistant strains of E. coli shortly after birth. The high prevalence of multidrug-resistant nonpathogenic E. coli in calves could be a significant source of resistance genes to other bacteria that share the same environment.
Figures
References
-
- Anderson, A. D., J. M. Nelson, S. Rossiter, and F. J. Angulo. 2003. Public health consequences of use of antimicrobial agents in food animals in the United States. Microb. Drug Resist. 9:373-379. - PubMed
-
- Arcangioli, M. A., S. Leroy-Setrin, J. L. Martel, and E. Chaslus-Dancla. 2000. Evolution of chloramphenicol resistance, with emergence of cross-resistance to florfenicol, in bovine Salmonella Typhimurium strains implicates definitive phage type (DT) 104. J. Med. Microbiol. 49:103-110. - PubMed
-
- Batchelor, M., F. A. Clifton-Hadley, A. D. Stallwood, G. A. Paiba, R. H. Davies, and E. Liebana. 2005. Detection of multiple cephalosporin-resistant Escherichia coli from a cattle fecal sample in Great Britain. Microb. Drug Resist. 11:58-61. - PubMed
-
- Berge, A. C. B., J. M. Adaska, and W. M. Sischo. 2004. Use of antibiotic susceptibility patterns and pulsed-field gel electrophoresis to compare historic and contemporary isolates of multi-drug resistant Salmonella enterica subsp. enterica serovar Newport. Appl. Environ. Microbiol. 70:318-323. - PMC - PubMed
-
- Bischoff, K. M., T. S. Edrington, T. R. Callaway, K. J. Genovese, and D. J. Nisbet. 2004. Characterization of antimicrobial resistant Salmonella Kinshasa from dairy calves in Texas. Lett. Appl. Microbiol. 38:140-145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical