Succession of bacterial communities during early plant development: transition from seed to root and effect of compost amendment
- PMID: 16751505
- PMCID: PMC1489615
- DOI: 10.1128/AEM.02771-05
Succession of bacterial communities during early plant development: transition from seed to root and effect of compost amendment
Abstract
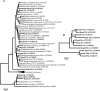
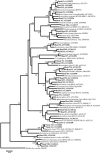
Compost amendments to soils and potting mixes are routinely applied to improve soil fertility and plant growth and health. These amendments, which contain high levels of organic matter and microbial cells, can influence microbial communities associated with plants grown in such soils. The purpose of this study was to follow the bacterial community compositions of seed and subsequent root surfaces in the presence and absence of compost in the potting mix. The bacterial community compositions of potting mixes, seed, and root surfaces sampled at three stages of plant growth were analyzed via general and newly developed Bacteroidetes-specific, PCR-denaturing gradient gel electrophoresis methodologies. These analyses revealed that seed surfaces were colonized primarily by populations detected in the initial potting mixes, many of which were not detected in subsequent root analyses. The most persistent bacterial populations detected in this study belonged to the genus Chryseobacterium (Bacteroidetes) and the family Oxalobacteraceae (Betaproteobacteria). The patterns of colonization by populations within these taxa differed significantly and may reflect differences in the physiology of these organisms. Overall, analyses of bacterial community composition revealed a surprising prevalence and diversity of Bacteroidetes in all treatments.
Figures




References
-
- Alfreider, A., S. Peters, C. C. Tebbe, A. Rangger, and H. Insam. 2002. Microbial community dynamics during composting of organic matter as determined by 16S ribosomal DNA analysis. Comp. Sci. Util. 10:303-312.
-
- Baudoin, E., E. Benizri, and A. Guckert. 2002. Impact of growth stage on the bacterial community structure along maize roots, as determined by metabolic and genetic fingerprinting. Appl. Soil Ecol. 19:135-145.
-
- Bazin, M. J., P. Markham, and E. M. Scott. 1990. Population dynamics and rhizosphere interactions, p. 99-127. In J. M. Lynch (ed.), The rhizosphere. John Wiley & Sons, Ltd., West Sussex, United Kingdom.
-
- Beffa, T., M. Blanc, L. Marilley, J. L. Fischer, P. F. Lyon, and M. Aragno. 1996. Taxonomic and metabolic microbial diversity during composting, p. 149-161. In M. De Bertoldi, P. Sequi, B. Lemmes, and T. Papi (ed.), The science of composting. M. Chapman and Hall, London, United Kingdom.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

