Microbial dioxygenase gene population shifts during polycyclic aromatic hydrocarbon biodegradation
- PMID: 16751518
- PMCID: PMC1489606
- DOI: 10.1128/AEM.02969-05
Microbial dioxygenase gene population shifts during polycyclic aromatic hydrocarbon biodegradation
Abstract
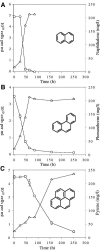
The degradation of polycyclic aromatic hydrocarbons (PAHs) by bacteria has been widely studied. While many pure cultures have been isolated and characterized for their ability to grow on PAHs, limited information is available on the diversity of microbes involved in PAH degradation in the environment. We have designed generic PCR primers targeting the gene fragment encoding the Rieske iron sulfur center common to all PAH dioxygenase enzymes. These Rieske primers were employed to track dioxygenase gene population shifts in soil enrichment cultures following exposure to naphthalene, phenanthrene, or pyrene. PAH degradation was monitored by gas chromatograph with flame ionization detection. DNA was extracted from the enrichment cultures following PAH degradation. 16S rRNA and Rieske gene fragments were PCR amplified from DNA extracted from each enrichment culture and an unamended treatment. The PCR products were cloned and sequenced. Molecular monitoring of the enrichment cultures before and after PAH degradation using denaturing gradient gel electrophoresis and 16S rRNA gene libraries suggests that specific phylotypes of bacteria were associated with the degradation of each PAH. Sequencing of the cloned Rieske gene fragments showed that different suites of genes were present in soil microbe populations under each enrichment culture condition. Many of the Rieske gene fragment sequences fell into clades which are distinct from the reference dioxygenase gene sequences used to design the PCR primers. The ability to profile not only the bacterial community but also the dioxygenases which they encode provides a powerful tool for both assessing bioremediation potential in the environment and for the discovery of novel dioxygenase genes.
Figures




References
-
- Ahn, Y., J. Sanseverino, and G. S. Sayler. 1999. Analyses of polycyclic aromatic hydrocarbon-degrading bacteria isolated from contaminated soils. Biodegradation 10:149-157. - PubMed
-
- Bodour, A. A., J. M. Wang, M. L. Brusseau, and R. M. Maier. 2003. Temporal change in culturable phenanthrene degraders in response to long-term exposure to phenanthrene in a soil column system. Environ. Microbiol. 5:888-895. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

